Lilach Barkat
PCMC-T1: Free-breathing myocardial T1 mapping with Physically-Constrained Motion Correction
Aug 22, 2023Abstract:T1 mapping is a quantitative magnetic resonance imaging (qMRI) technique that has emerged as a valuable tool in the diagnosis of diffuse myocardial diseases. However, prevailing approaches have relied heavily on breath-hold sequences to eliminate respiratory motion artifacts. This limitation hinders accessibility and effectiveness for patients who cannot tolerate breath-holding. Image registration can be used to enable free-breathing T1 mapping. Yet, inherent intensity differences between the different time points make the registration task challenging. We introduce PCMC-T1, a physically-constrained deep-learning model for motion correction in free-breathing T1 mapping. We incorporate the signal decay model into the network architecture to encourage physically-plausible deformations along the longitudinal relaxation axis. We compared PCMC-T1 to baseline deep-learning-based image registration approaches using a 5-fold experimental setup on a publicly available dataset of 210 patients. PCMC-T1 demonstrated superior model fitting quality (R2: 0.955) and achieved the highest clinical impact (clinical score: 3.93) compared to baseline methods (0.941, 0.946 and 3.34, 3.62 respectively). Anatomical alignment results were comparable (Dice score: 0.9835 vs. 0.984, 0.988). Our code and trained models are available at https://github.com/eyalhana/PCMC-T1.
Image translation of Ultrasound to Pseudo Anatomical Display Using Artificial Intelligence
Feb 16, 2022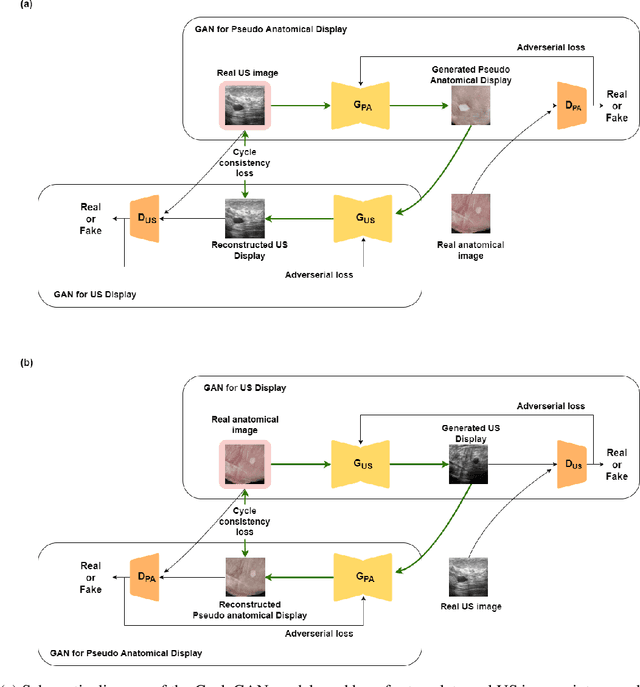
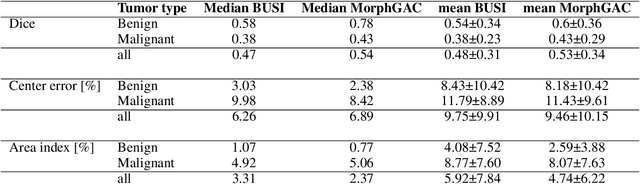
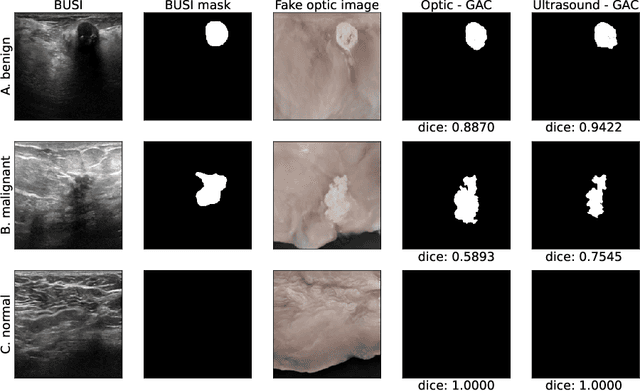
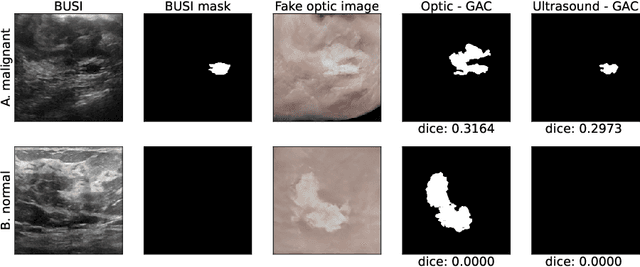
Abstract:Ultrasound is the second most used modality in medical imaging. It is cost effective, hazardless, portable and implemented routinely in numerous clinical procedures. Nonetheless, image quality is characterized by granulated appearance, poor SNR and speckle noise. Specific for malignant tumors, the margins are blurred and indistinct. Thus, there is a great need for improving ultrasound image quality. We hypothesize that this can be achieved by translation into a more realistic anatomic display, using neural networks. In order to achieve this goal, the preferable approach would be to use a set of paired images. However, this is practically impossible in our case. Therefore, CycleGAN was used, to learn each domain properties separately and enforce cross domain cycle consistency. The two datasets which were used for training the model were "Breast Ultrasound Images" (BUSI) and a set of optic images of poultry breast tissue samples acquired at our lab. The generated pseudo anatomical images provide improved visual discrimination of the lesions with clearer border definition and pronounced contrast. Furthermore, the algorithm manages to overcome the acoustic shadows artifacts commonly appearing in ultrasonic images. In order to evaluate the preservation of the anatomical features, the lesions in the ultrasonic images and the generated pseudo anatomical images were both automatically segmented and compared. This comparison yielded median dice score of 0.78 for the benign tumors and 0.43 for the malignancies. Median lesion center error of 2.38% and 8.42% for the benign and malignancies respectively and median area error index of 0.77% and 5.06% for the benign and malignancies respectively. In conclusion, these generated pseudo anatomical images, which are presented in a more intuitive way, preserve tissue anatomy and can potentially simplify the diagnosis and improve the clinical outcome.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge