Leigh H. Whitehead
Submanifold Sparse Convolutional Networks for Automated 3D Segmentation of Kidneys and Kidney Tumours in Computed Tomography
Nov 06, 2025Abstract:The accurate delineation of tumours in radiological images like Computed Tomography is a very specialised and time-consuming task, and currently a bottleneck preventing quantitative analyses to be performed routinely in the clinical setting. For this reason, developing methods for the automated segmentation of tumours in medical imaging is of the utmost importance and has driven significant efforts in recent years. However, challenges regarding the impracticality of 3D scans, given the large amount of voxels to be analysed, usually requires the downsampling of such images or using patches thereof when applying traditional convolutional neural networks. To overcome this problem, in this paper we propose a new methodology that uses, divided into two stages, voxel sparsification and submanifold sparse convolutional networks. This method allows segmentations to be performed with high-resolution inputs and a native 3D model architecture, obtaining state-of-the-art accuracies while significantly reducing the computational resources needed in terms of GPU memory and time. We studied the deployment of this methodology in the context of Computed Tomography images of renal cancer patients from the KiTS23 challenge, and our method achieved results competitive with the challenge winners, with Dice similarity coefficients of 95.8% for kidneys + masses, 85.7% for tumours + cysts, and 80.3% for tumours alone. Crucially, our method also offers significant computational improvements, achieving up to a 60% reduction in inference time and up to a 75\% reduction in VRAM usage compared to an equivalent dense architecture, across both CPU and various GPU cards tested.
Automated Segmentation of Computed Tomography Images with Submanifold Sparse Convolutional Networks
Dec 06, 2022



Abstract:Quantitative cancer image analysis relies on the accurate delineation of tumours, a very specialised and time-consuming task. For this reason, methods for automated segmentation of tumours in medical imaging have been extensively developed in recent years, being Computed Tomography one of the most popular imaging modalities explored. However, the large amount of 3D voxels in a typical scan is prohibitive for the entire volume to be analysed at once in conventional hardware. To overcome this issue, the processes of downsampling and/or resampling are generally implemented when using traditional convolutional neural networks in medical imaging. In this paper, we propose a new methodology that introduces a process of sparsification of the input images and submanifold sparse convolutional networks as an alternative to downsampling. As a proof of concept, we applied this new methodology to Computed Tomography images of renal cancer patients, obtaining performances of segmentations of kidneys and tumours competitive with previous methods (~84.6% Dice similarity coefficient), while achieving a significant improvement in computation time (2-3 min per training epoch).
Image-based model parameter optimisation using Model-Assisted Generative Adversarial Networks
Nov 30, 2018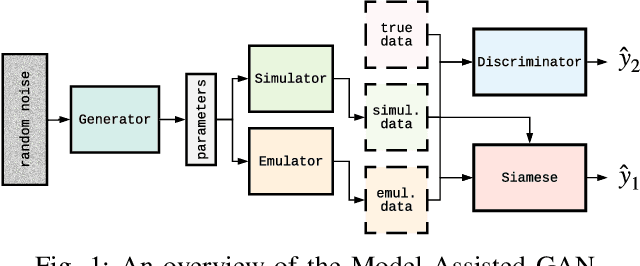
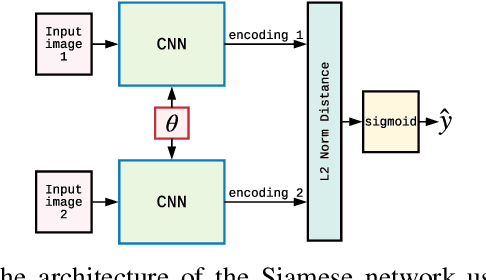
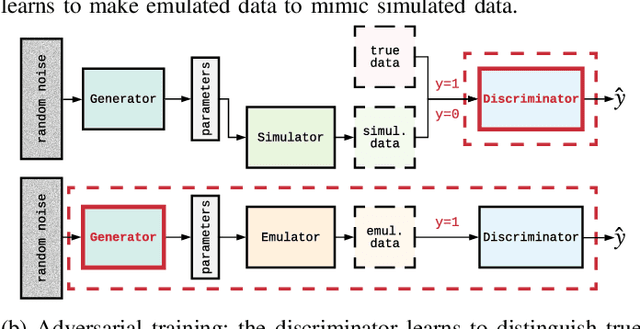
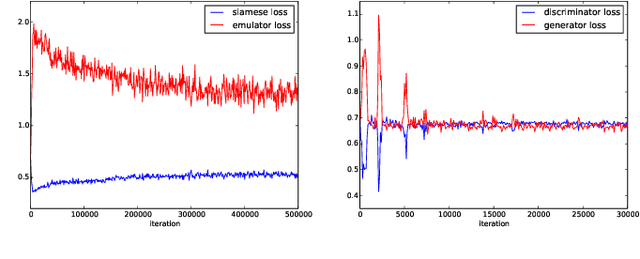
Abstract:We propose and demonstrate the use of a Model-Assisted Generative Adversarial Network to produce simulated images that accurately match true images through the variation of underlying model parameters that describe the image generation process. The generator learns the parameter values that give images that best match the true images. Two case studies show the excellent agreement between the generated best match parameters and the true parameters. The best match parameter values that produce the most accurate simulated images can be extracted and used to re-tune the default simulation to minimise any bias when applying image recognition techniques to simulated and true images. In the case of a real-world experiment, the true data is replaced by experimental data with unknown true parameter values. The Model-Assisted Generative Adversarial Network uses a convolutional neural network to emulate the simulation for all parameter values that, when trained, can be used as a conditional generator for fast image production.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge