Kim Steenstrup Pedersen
A Time Series Dataset of NIR Spectra and RGB and NIR-HSI Images of the Barley Germination Process
Apr 23, 2025Abstract:We provide an open-source dataset of RGB and NIR-HSI (near-infrared hyperspectral imaging) images with associated segmentation masks and NIR spectra of 2242 individual malting barley kernels. We imaged every kernel pre-exposure to moisture and every 24 hours after exposure to moisture for five consecutive days. Every barley kernel was labeled as germinated or not germinated during each image acquisition. The barley kernels were imaged with black filter paper as the background, facilitating straight-forward intensity threshold-based segmentation, e.g., by Otsu's method. This dataset facilitates time series analysis of germination time for barley kernels using either RGB image analysis, NIR spectral analysis, NIR-HSI analysis, or a combination hereof.
Transforming hyperspectral images into chemical maps: A new deep learning based approach to hyperspectral image processing
Apr 19, 2025Abstract:Current approaches to chemical map generation from hyperspectral images are based on models such as partial least squares (PLS) regression, generating pixel-wise predictions that do not consider spatial context and suffer from a high degree of noise. This study proposes an end-to-end deep learning approach using a modified version of U-Net and a custom loss function to directly obtain chemical maps from hyperspectral images, skipping all intermediate steps required for traditional pixel-wise analysis. We compare the U-Net with the traditional PLS regression on a real dataset of pork belly samples with associated mean fat reference values. The U-Net obtains a test set root mean squared error of between 9% and 13% lower than that of PLS regression on the task of mean fat prediction. At the same time, U-Net generates fine detail chemical maps where 99.91% of the variance is spatially correlated. Conversely, only 2.53% of the variance in the PLS-generated chemical maps is spatially correlated, indicating that each pixel-wise prediction is largely independent of neighboring pixels. Additionally, while the PLS-generated chemical maps contain predictions far beyond the physically possible range of 0-100%, U-Net learns to stay inside this range. Thus, the findings of this study indicate that U-Net is superior to PLS for chemical map generation.
The Phantom of the Elytra -- Phylogenetic Trait Extraction from Images of Rove Beetles Using Deep Learning -- Is the Mask Enough?
Feb 06, 2025Abstract:Phylogenetic analysis traditionally relies on labor-intensive manual extraction of morphological traits, limiting its scalability for large datasets. Recent advances in deep learning offer the potential to automate this process, but the effectiveness of different morphological representations for phylogenetic trait extraction remains poorly understood. In this study, we compare the performance of deep learning models using three distinct morphological representations - full segmentations, binary masks, and Fourier descriptors of beetle outlines. We test this on the Rove-Tree-11 dataset, a curated collection of images from 215 rove beetle species. Our results demonstrate that the mask-based model outperformed the others, achieving a normalized Align Score of 0.33 plus/minus 0.02 on the test set, compared to 0.45 plus/minus 0.01 for the Fourier-based model and 0.39 plus/minus 0.07 for the segmentation-based model. The performance of the mask-based model likely reflects its ability to capture shape features while taking advantage of the depth and capacity of the ResNet50 architecture. These results also indicate that dorsal textural features, at least in this group of beetles, may be of lowered phylogenetic relevance, though further investigation is necessary to confirm this. In contrast, the Fourier-based model suffered from reduced capacity and occasional inaccuracies in outline approximations, particularly in fine structures like legs. These findings highlight the importance of selecting appropriate morphological representations for automated phylogenetic studies and the need for further research into explainability in automatic morphological trait extraction.
Analyzing Near-Infrared Hyperspectral Imaging for Protein Content Regression and Grain Variety Classification Using Bulk References and Varying Grain-to-Background Ratios
Nov 07, 2023Abstract:Based on previous work, we assess the use of NIR-HSI images for calibrating models on two datasets, focusing on protein content regression and grain variety classification. Limited reference data for protein content is expanded by subsampling and associating it with the bulk sample. However, this method introduces significant biases due to skewed leptokurtic prediction distributions, affecting both PLS-R and deep CNN models. We propose adjustments to mitigate these biases, improving mean protein reference predictions. Additionally, we investigate the impact of grain-to-background ratios on both tasks. Higher ratios yield more accurate predictions, but including lower-ratio images in calibration enhances model robustness for such scenarios.
Sacrificing information for the greater good: how to select photometric bands for optimal accuracy
Jul 06, 2016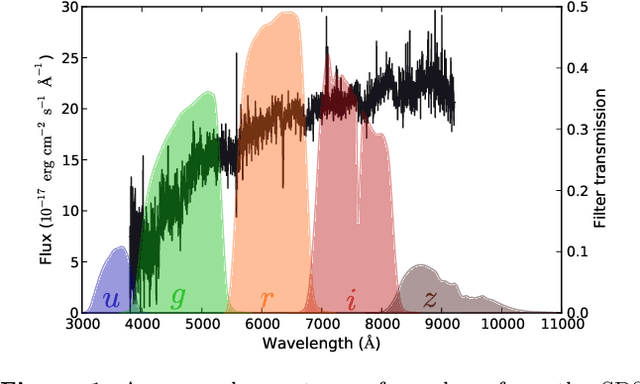
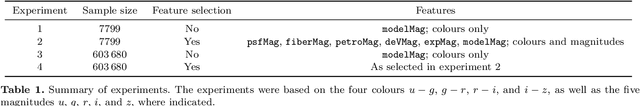
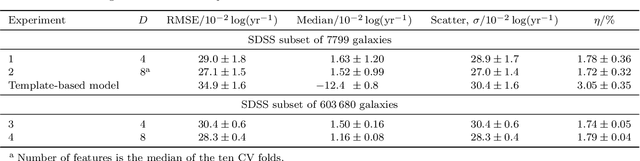
Abstract:Large-scale surveys make huge amounts of photometric data available. Because of the sheer amount of objects, spectral data cannot be obtained for all of them. Therefore it is important to devise techniques for reliably estimating physical properties of objects from photometric information alone. These estimates are needed to automatically identify interesting objects worth a follow-up investigation as well as to produce the required data for a statistical analysis of the space covered by a survey. We argue that machine learning techniques are suitable to compute these estimates accurately and efficiently. This study promotes a feature selection algorithm, which selects the most informative magnitudes and colours for a given task of estimating physical quantities from photometric data alone. Using k nearest neighbours regression, a well-known non-parametric machine learning method, we show that using the found features significantly increases the accuracy of the estimations compared to using standard features and standard methods. We illustrate the usefulness of the approach by estimating specific star formation rates (sSFRs) and redshifts (photo-z's) using only the broad-band photometry from the Sloan Digital Sky Survey (SDSS). For estimating sSFRs, we demonstrate that our method produces better estimates than traditional spectral energy distribution (SED) fitting. For estimating photo-z's, we show that our method produces more accurate photo-z's than the method employed by SDSS. The study highlights the general importance of performing proper model selection to improve the results of machine learning systems and how feature selection can provide insights into the predictive relevance of particular input features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge