Kenneth A. Iczkowski
Department of Pathology, Medical College of Wisconsin, Milwaukee, WI, USA
Label-free evaluation of lung and heart transplant biopsies using virtual staining
Sep 09, 2024



Abstract:Organ transplantation serves as the primary therapeutic strategy for end-stage organ failures. However, allograft rejection is a common complication of organ transplantation. Histological assessment is essential for the timely detection and diagnosis of transplant rejection and remains the gold standard. Nevertheless, the traditional histochemical staining process is time-consuming, costly, and labor-intensive. Here, we present a panel of virtual staining neural networks for lung and heart transplant biopsies, which digitally convert autofluorescence microscopic images of label-free tissue sections into their brightfield histologically stained counterparts, bypassing the traditional histochemical staining process. Specifically, we virtually generated Hematoxylin and Eosin (H&E), Masson's Trichrome (MT), and Elastic Verhoeff-Van Gieson (EVG) stains for label-free transplant lung tissue, along with H&E and MT stains for label-free transplant heart tissue. Subsequent blind evaluations conducted by three board-certified pathologists have confirmed that the virtual staining networks consistently produce high-quality histology images with high color uniformity, closely resembling their well-stained histochemical counterparts across various tissue features. The use of virtually stained images for the evaluation of transplant biopsies achieved comparable diagnostic outcomes to those obtained via traditional histochemical staining, with a concordance rate of 82.4% for lung samples and 91.7% for heart samples. Moreover, virtual staining models create multiple stains from the same autofluorescence input, eliminating structural mismatches observed between adjacent sections stained in the traditional workflow, while also saving tissue, expert time, and staining costs.
Pathologist-Level Grading of Prostate Biopsies with Artificial Intelligence
Jul 02, 2019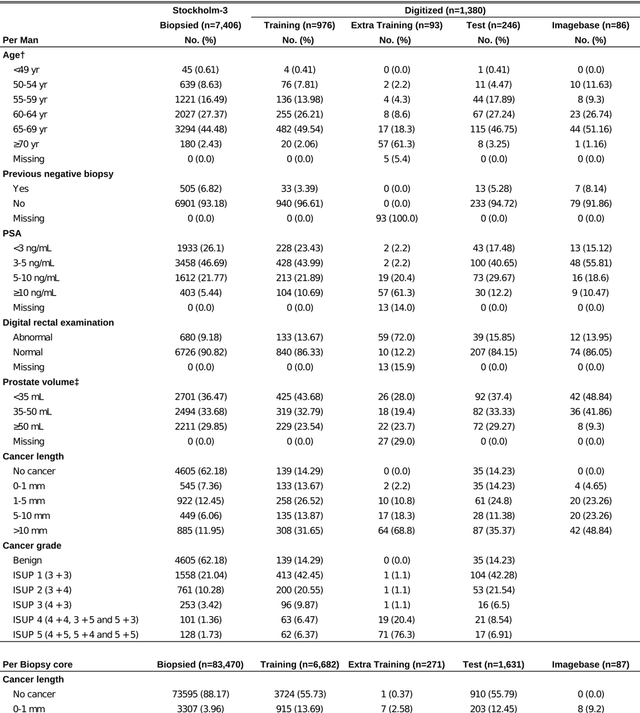
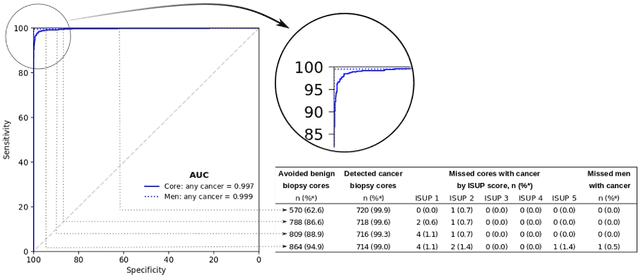
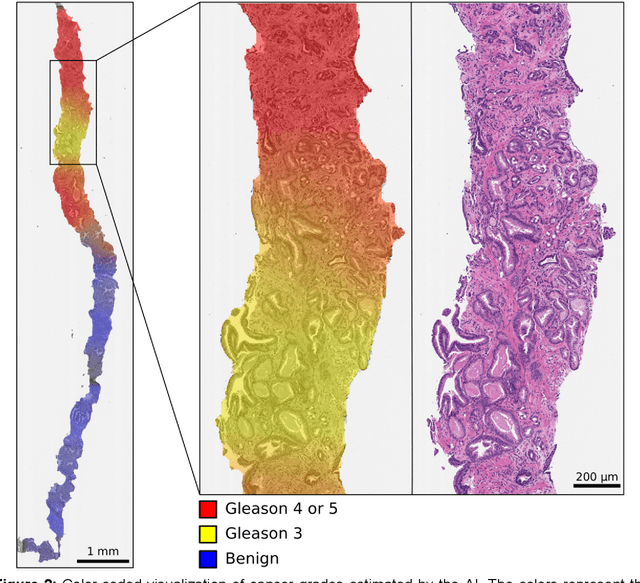
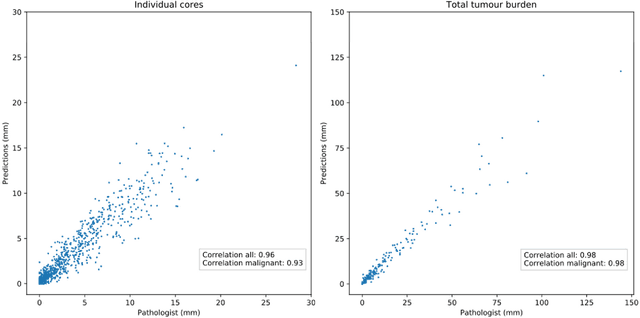
Abstract:Background: An increasing volume of prostate biopsies and a world-wide shortage of uro-pathologists puts a strain on pathology departments. Additionally, the high intra- and inter-observer variability in grading can result in over- and undertreatment of prostate cancer. Artificial intelligence (AI) methods may alleviate these problems by assisting pathologists to reduce workload and harmonize grading. Methods: We digitized 6,682 needle biopsies from 976 participants in the population based STHLM3 diagnostic study to train deep neural networks for assessing prostate biopsies. The networks were evaluated by predicting the presence, extent, and Gleason grade of malignant tissue for an independent test set comprising 1,631 biopsies from 245 men. We additionally evaluated grading performance on 87 biopsies individually graded by 23 experienced urological pathologists from the International Society of Urological Pathology. We assessed discriminatory performance by receiver operating characteristics (ROC) and tumor extent predictions by correlating predicted millimeter cancer length against measurements by the reporting pathologist. We quantified the concordance between grades assigned by the AI and the expert urological pathologists using Cohen's kappa. Results: The performance of the AI to detect and grade cancer in prostate needle biopsy samples was comparable to that of international experts in prostate pathology. The AI achieved an area under the ROC curve of 0.997 for distinguishing between benign and malignant biopsy cores, and 0.999 for distinguishing between men with or without prostate cancer. The correlation between millimeter cancer predicted by the AI and assigned by the reporting pathologist was 0.96. For assigning Gleason grades, the AI achieved an average pairwise kappa of 0.62. This was within the range of the corresponding values for the expert pathologists (0.60 to 0.73).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge