Joachim Behar
Extraction of clinical information from the non-invasive fetal electrocardiogram
May 27, 2016
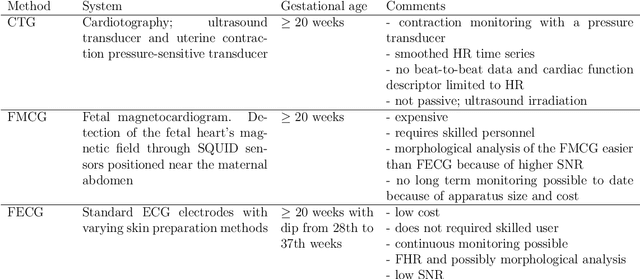
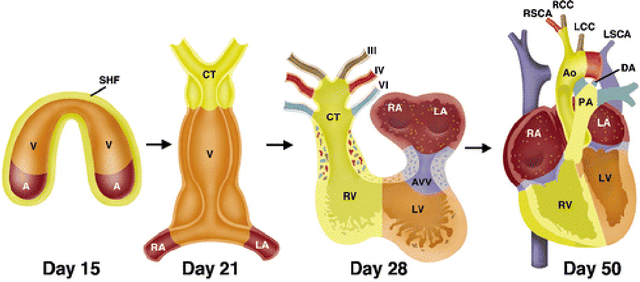
Abstract:Estimation of the fetal heart rate (FHR) has gained interest in the last century, low heart rate variability has been studied to identify intrauterine growth restricted fetuses (prepartum), and abnormal FHR patterns have been associated with fetal distress during delivery (intrapartum). Several monitoring techniques have been proposed for FHR estimation, including auscultation and Doppler ultrasound. This thesis focuses on the extraction of the non-invasive fetal electrocardiogram (NI-FECG) recorded from a limited set of abdominal sensors. The main challenge with NI-FECG extraction techniques is the low signal-to-noise ratio of the FECG signal on the abdominal mixture signal which consists of a dominant maternal ECG component, FECG and noise. However the NI-FECG offers many advantages over the alternative fetal monitoring techniques, the most important one being the opportunity to enable morphological analysis of the FECG which is vital for determining whether an observed FHR event is normal or pathological. In order to advance the field of NI-FECG signal processing, the development of standardised public databases and benchmarking of a number of published and novel algorithms was necessary.
Fusing Continuous-valued Medical Labels using a Bayesian Model
Jun 13, 2015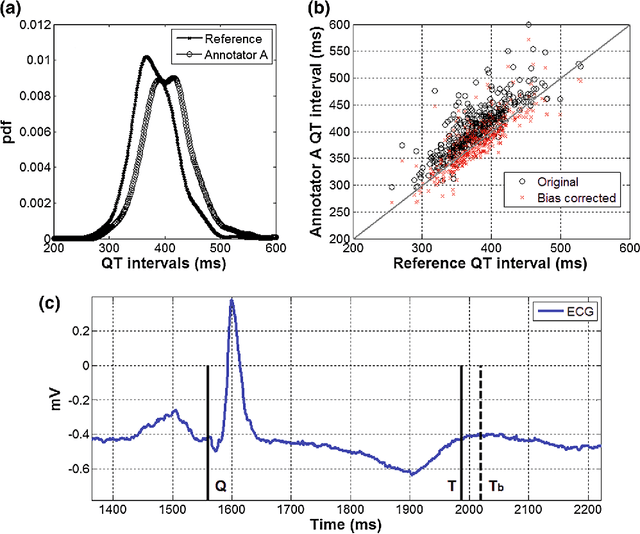
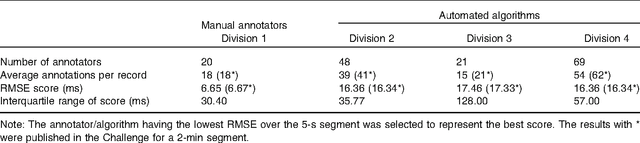
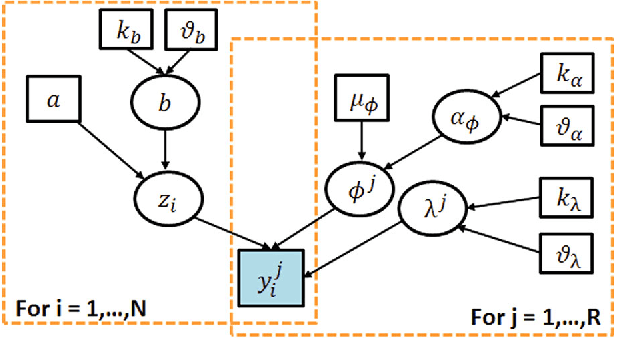
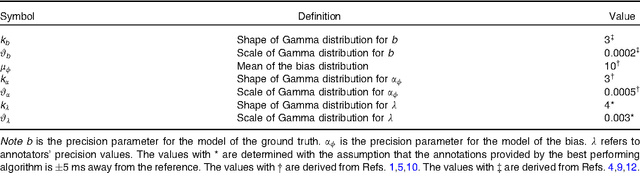
Abstract:With the rapid increase in volume of time series medical data available through wearable devices, there is a need to employ automated algorithms to label data. Examples of labels include interventions, changes in activity (e.g. sleep) and changes in physiology (e.g. arrhythmias). However, automated algorithms tend to be unreliable resulting in lower quality care. Expert annotations are scarce, expensive, and prone to significant inter- and intra-observer variance. To address these problems, a Bayesian Continuous-valued Label Aggregator(BCLA) is proposed to provide a reliable estimation of label aggregation while accurately infer the precision and bias of each algorithm. The BCLA was applied to QT interval (pro-arrhythmic indicator) estimation from the electrocardiogram using labels from the 2006 PhysioNet/Computing in Cardiology Challenge database. It was compared to the mean, median, and a previously proposed Expectation Maximization (EM) label aggregation approaches. While accurately predicting each labelling algorithm's bias and precision, the root-mean-square error of the BCLA was 11.78$\pm$0.63ms, significantly outperforming the best Challenge entry (15.37$\pm$2.13ms) as well as the EM, mean, and median voting strategies (14.76$\pm$0.52ms, 17.61$\pm$0.55ms, and 14.43$\pm$0.57ms respectively with $p<0.0001$).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge