Jan Moltz
Constraining Volume Change in Learned Image Registration for Lung CTs
Nov 29, 2020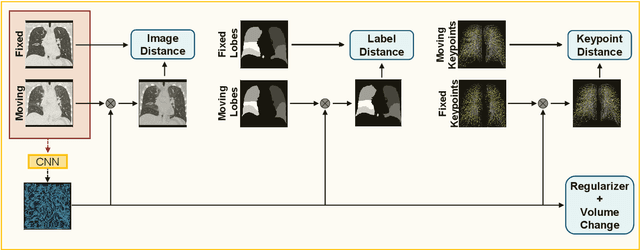
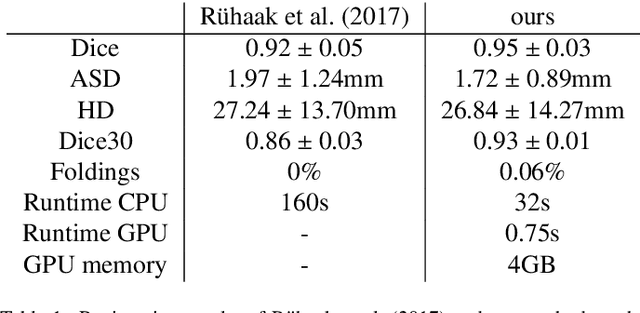
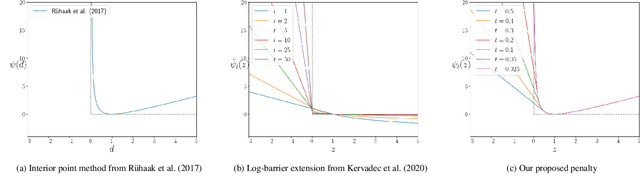

Abstract:Deep-learning-based registration methods emerged as a fast alternative to conventional registration methods. However, these methods often still cannot achieve the same performance as conventional registration methods, because they are either limited to small deformation or they fail to handle a superposition of large and small deformations without producing implausible deformation fields with foldings inside. In this paper, we identify important strategies of conventional registration methods for lung registration and successfully developed the deep-learning counterpart. We employ a Gaussian-pyramid-based multilevel framework that can solve the image registration optimization in a coarse-to-fine fashion. Furthermore, we prevent foldings of the deformation field and restrict the determinant of the Jacobian to physiologically meaningful values by combining a volume change penalty with a curvature regularizer in the loss function. Keypoint correspondences are integrated to focus on the alignment of smaller structures. We perform an extensive evaluation to assess the accuracy, the robustness, the plausibility of the estimated deformation fields, and the transferability of our registration approach. We show that it archives state-of-the-art results on the COPDGene dataset compared to the challenge winning conventional registration method with much shorter execution time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge