H. R. Tizhoosh
Kimia Lab, University of Waterloo, Waterloo, ON, Canada, Vector Institute, MaRS Centre, Toronto, Canada
The Effects of Image Pre- and Post-Processing, Wavelet Decomposition, and Local Binary Patterns on U-Nets for Skin Lesion Segmentation
Apr 30, 2018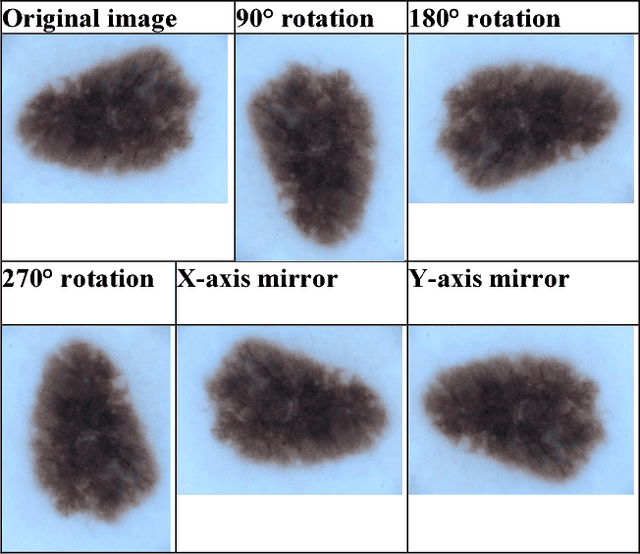
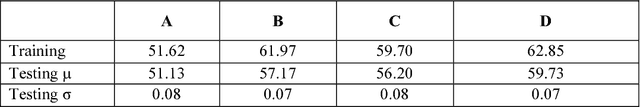
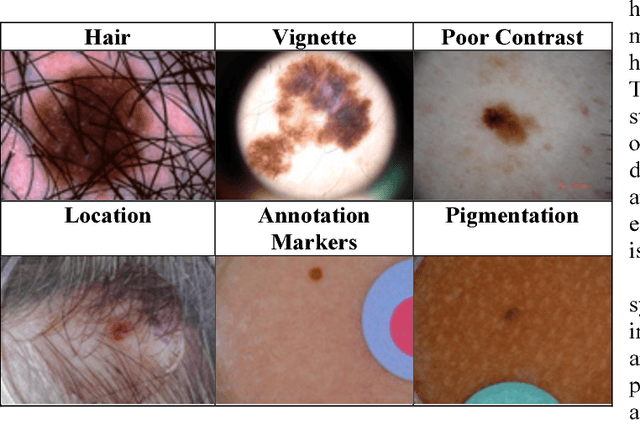

Abstract:Skin cancer is a widespread, global, and potentially deadly disease, which over the last three decades has afflicted more lives in the USA than all other forms of cancer combined. There have been a lot of promising recent works utilizing deep network architectures, such as FCNs, U-Nets, and ResNets, for developing automated skin lesion segmentation. This paper investigates various pre- and post-processing techniques for improving the performance of U-Nets as measured by the Jaccard Index. The dataset provided as part of the "2017 ISBI Challenges on Skin Lesion Analysis Towards Melanoma Detection" was used for this evaluation and the performance of the finalist competitors was the standard for comparison. The pre-processing techniques employed in the proposed system included contrast enhancement, artifact removal, and vignette correction. More advanced image transformations, such as local binary patterns and wavelet decomposition, were also employed to augment the raw grayscale images used as network input features. While the performance of the proposed system fell short of the winners of the challenge, it was determined that using wavelet decomposition as an early transformation step improved the overall performance of the system over pre- and post-processing steps alone.
Local Radon Descriptors for Image Search
Oct 11, 2017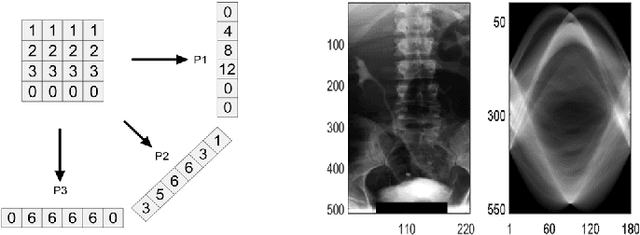
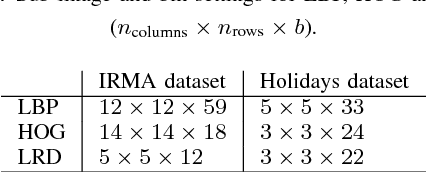
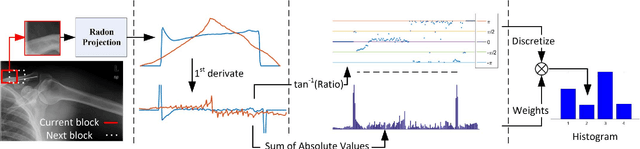
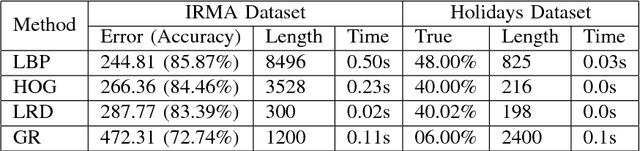
Abstract:Radon transform and its inverse operation are important techniques in medical imaging tasks. Recently, there has been renewed interest in Radon transform for applications such as content-based medical image retrieval. However, all studies so far have used Radon transform as a global or quasi-global image descriptor by extracting projections of the whole image or large sub-images. This paper attempts to show that the dense sampling to generate the histogram of local Radon projections has a much higher discrimination capability than the global one. In this paper, we introduce Local Radon Descriptor (LRD) and apply it to the IRMA dataset, which contains 14,410 x-ray images as well as to the INRIA Holidays dataset with 1,990 images. Our results show significant improvement in retrieval performance by using LRD versus its global version. We also demonstrate that LRD can deliver results comparable to well-established descriptors like LBP and HOG.
Convolutional Neural Networks for Histopathology Image Classification: Training vs. Using Pre-Trained Networks
Oct 11, 2017

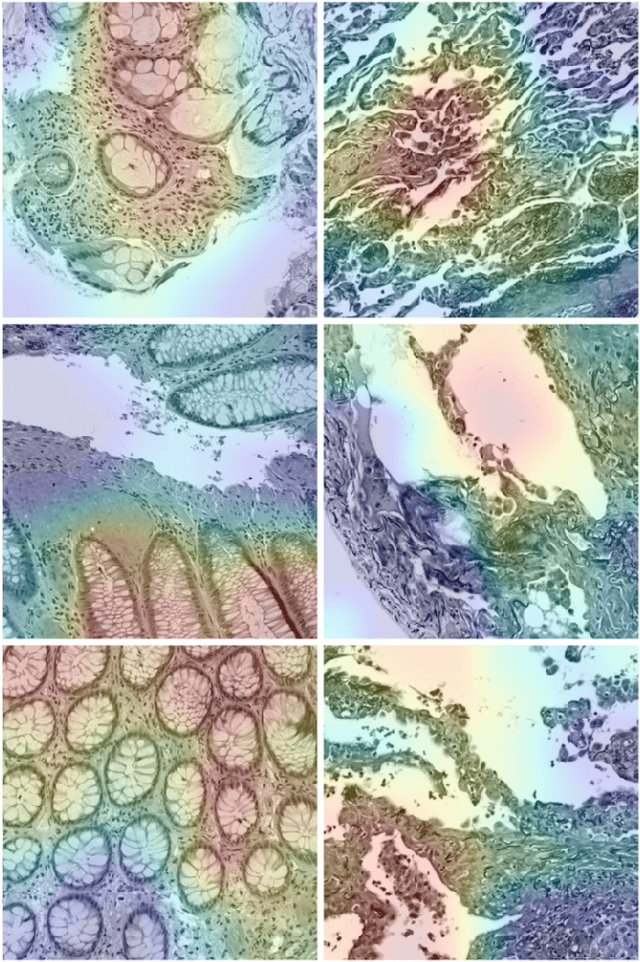
Abstract:We explore the problem of classification within a medical image data-set based on a feature vector extracted from the deepest layer of pre-trained Convolution Neural Networks. We have used feature vectors from several pre-trained structures, including networks with/without transfer learning to evaluate the performance of pre-trained deep features versus CNNs which have been trained by that specific dataset as well as the impact of transfer learning with a small number of samples. All experiments are done on Kimia Path24 dataset which consists of 27,055 histopathology training patches in 24 tissue texture classes along with 1,325 test patches for evaluation. The result shows that pre-trained networks are quite competitive against training from scratch. As well, fine-tuning does not seem to add any tangible improvement for VGG16 to justify additional training while we observed considerable improvement in retrieval and classification accuracy when we fine-tuned the Inception structure.
Fast Barcode Retrieval for Consensus Contouring
Sep 28, 2017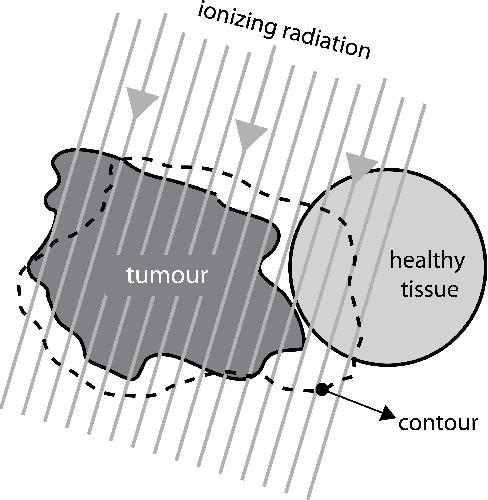
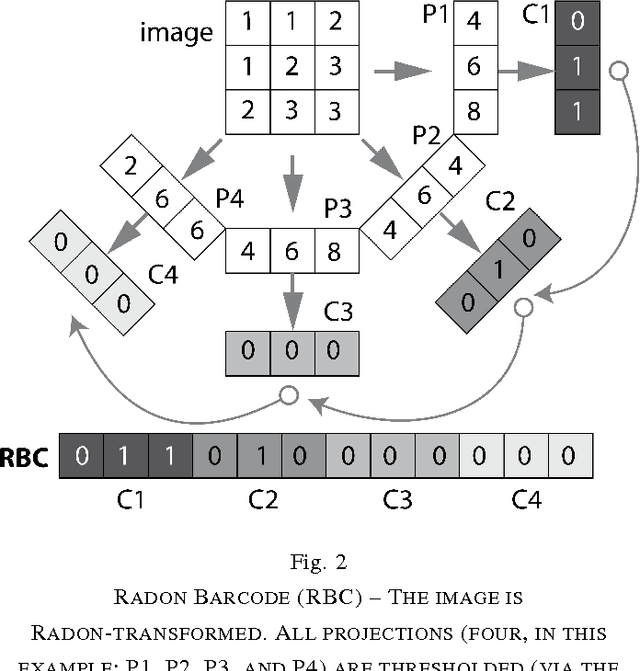
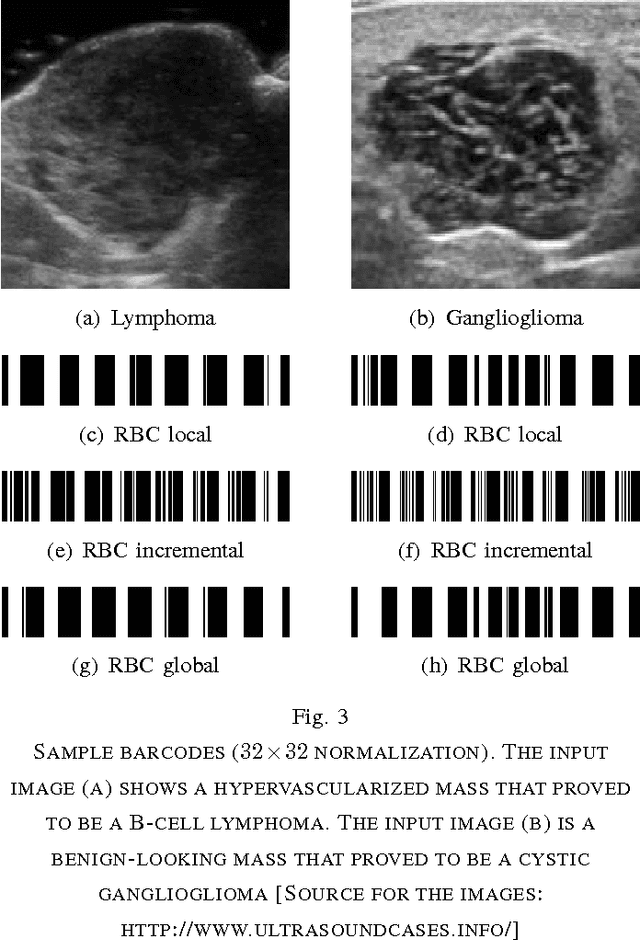
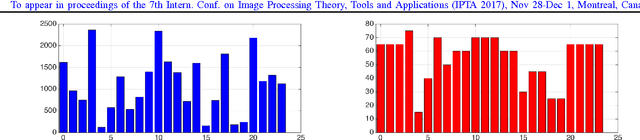
Abstract:Marking tumors and organs is a challenging task suffering from both inter- and intra-observer variability. The literature quantifies observer variability by generating consensus among multiple experts when they mark the same image. Automatically building consensus contours to establish quality assurance for image segmentation is presently absent in the clinical practice. As the \emph{big data} becomes more and more available, techniques to access a large number of existing segments of multiple experts becomes possible. Fast algorithms are, hence, required to facilitate the search for similar cases. The present work puts forward a potential framework that tested with small datasets (both synthetic and real images) displays the reliability of finding similar images. In this paper, the idea of content-based barcodes is used to retrieve similar cases in order to build consensus contours in medical image segmentation. This approach may be regarded as an extension of the conventional atlas-based segmentation that generally works with rather small atlases due to required computational expenses. The fast segment-retrieval process via barcodes makes it possible to create and use large atlases, something that directly contributes to the quality of the consensus building. Because the accuracy of experts' contours must be measured, we first used 500 synthetic prostate images with their gold markers and delineations by 20 simulated users. The fast barcode-guided computed consensus delivered an average error of $8\%\!\pm\!5\%$ compared against the gold standard segments. Furthermore, we used magnetic resonance images of prostates from 15 patients delineated by 5 oncologists and selected the best delineations to serve as the gold-standard segments. The proposed barcode atlas achieved a Jaccard overlap of $87\%\!\pm\!9\%$ with the contours of the gold-standard segments.
A Comparative Study of CNN, BoVW and LBP for Classification of Histopathological Images
Sep 27, 2017
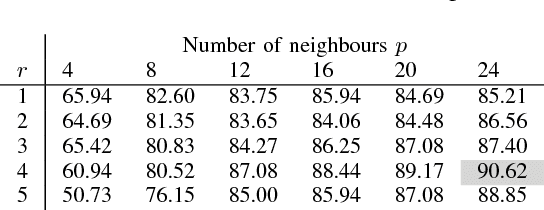
Abstract:Despite the progress made in the field of medical imaging, it remains a large area of open research, especially due to the variety of imaging modalities and disease-specific characteristics. This paper is a comparative study describing the potential of using local binary patterns (LBP), deep features and the bag-of-visual words (BoVW) scheme for the classification of histopathological images. We introduce a new dataset, \emph{KIMIA Path960}, that contains 960 histopathology images belonging to 20 different classes (different tissue types). We make this dataset publicly available. The small size of the dataset and its inter- and intra-class variability makes it ideal for initial investigations when comparing image descriptors for search and classification in complex medical imaging cases like histopathology. We investigate deep features, LBP histograms and BoVW to classify the images via leave-one-out validation. The accuracy of image classification obtained using LBP was 90.62\% while the highest accuracy using deep features reached 94.72\%. The dictionary approach (BoVW) achieved 96.50\%. Deep solutions may be able to deliver higher accuracies but they need extensive training with a large number of (balanced) image datasets.
Combining Real-Valued and Binary Gabor-Radon Features for Classification and Search in Medical Imaging Archives
Sep 27, 2017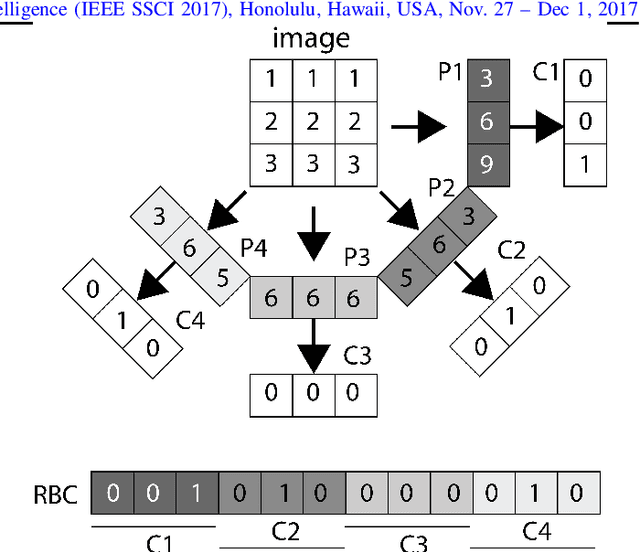
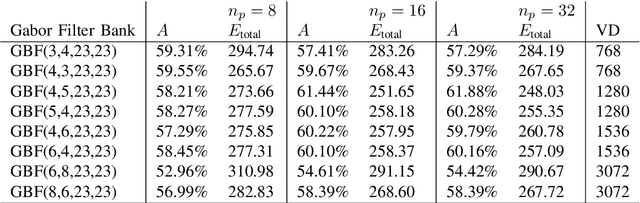
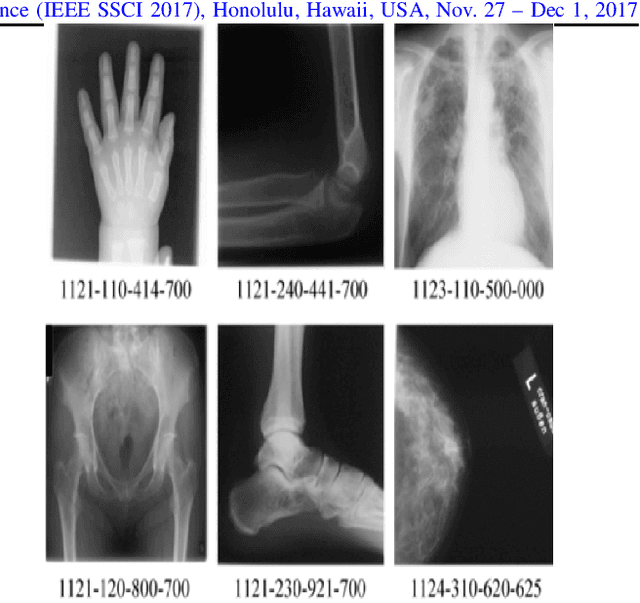
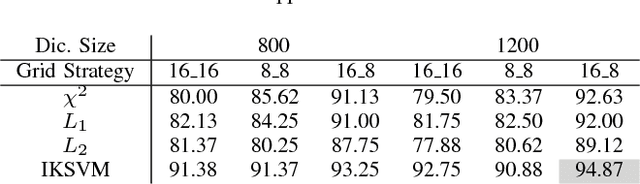
Abstract:Content-based image retrieval (CBIR) of medical images in large datasets to identify similar images when a query image is given can be very useful in improving the diagnostic decision of the clinical experts and as well in educational scenarios. In this paper, we used two stage classification and retrieval approach to retrieve similar images. First, the Gabor filters are applied to Radon-transformed images to extract features and to train a multi-class SVM. Then based on the classification results and using an extracted Gabor barcode, similar images are retrieved. The proposed method was tested on IRMA dataset which contains more than 14,000 images. Experimental results show the efficiency of our approach in retrieving similar images compared to other Gabor-Radon-oriented methods.
Skin Lesion Segmentation: U-Nets versus Clustering
Sep 27, 2017


Abstract:Many automatic skin lesion diagnosis systems use segmentation as a preprocessing step to diagnose skin conditions because skin lesion shape, border irregularity, and size can influence the likelihood of malignancy. This paper presents, examines and compares two different approaches to skin lesion segmentation. The first approach uses U-Nets and introduces a histogram equalization based preprocessing step. The second approach is a C-Means clustering based approach that is much simpler to implement and faster to execute. The Jaccard Index between the algorithm output and hand segmented images by dermatologists is used to evaluate the proposed algorithms. While many recently proposed deep neural networks to segment skin lesions require a significant amount of computational power for training (i.e., computer with GPUs), the main objective of this paper is to present methods that can be used with only a CPU. This severely limits, for example, the number of training instances that can be presented to the U-Net. Comparing the two proposed algorithms, U-Nets achieved a significantly higher Jaccard Index compared to the clustering approach. Moreover, using the histogram equalization for preprocessing step significantly improved the U-Net segmentation results.
Learning Autoencoded Radon Projections
Sep 27, 2017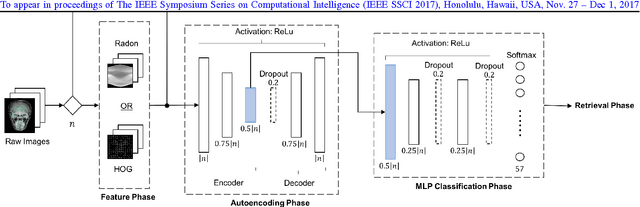
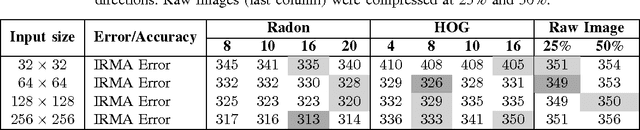
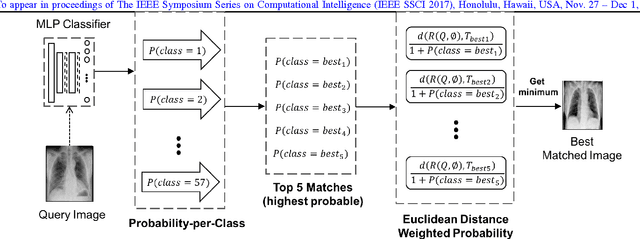
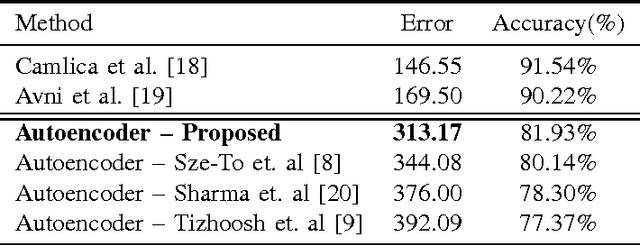
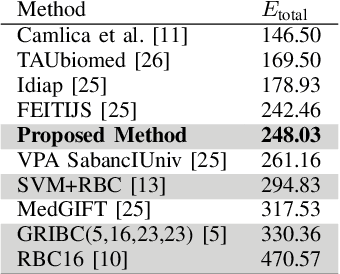
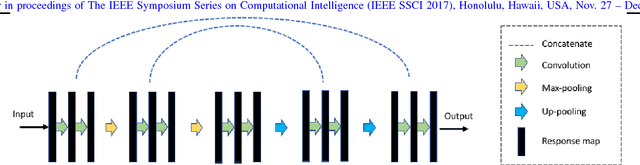
Abstract:Autoencoders have been recently used for encoding medical images. In this study, we design and validate a new framework for retrieving medical images by classifying Radon projections, compressed in the deepest layer of an autoencoder. As the autoencoder reduces the dimensionality, a multilayer perceptron (MLP) can be employed to classify the images. The integration of MLP promotes a rather shallow learning architecture which makes the training faster. We conducted a comparative study to examine the capabilities of autoencoders for different inputs such as raw images, Histogram of Oriented Gradients (HOG) and normalized Radon projections. Our framework is benchmarked on IRMA dataset containing $14,410$ x-ray images distributed across $57$ different classes. Experiments show an IRMA error of $313$ (equivalent to $\approx 82\%$ accuracy) outperforming state-of-the-art works on retrieval from IRMA dataset using autoencoders.
Classification and Retrieval of Digital Pathology Scans: A New Dataset
May 22, 2017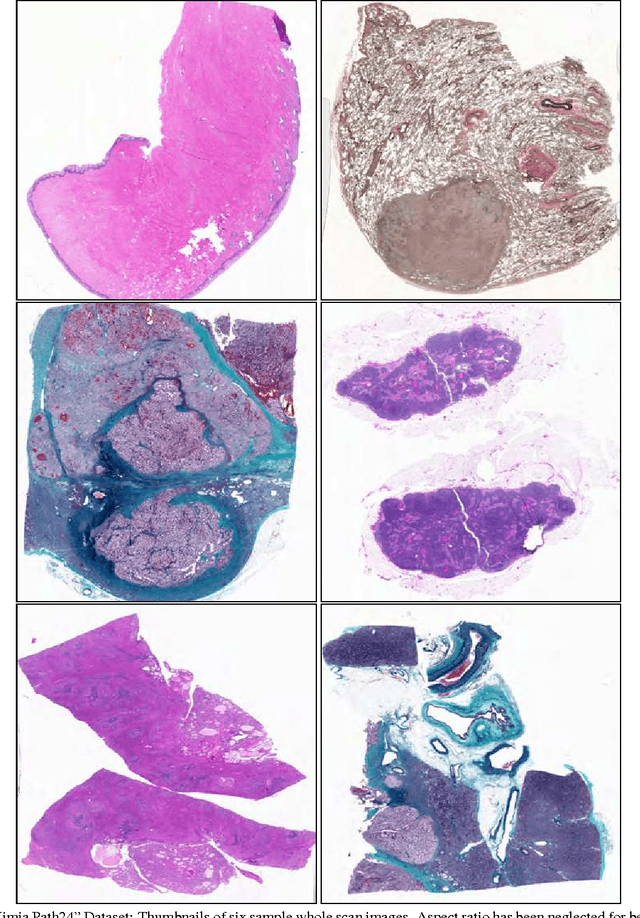


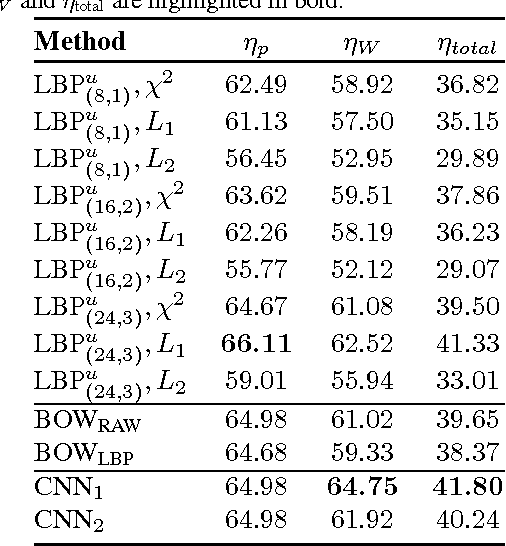
Abstract:In this paper, we introduce a new dataset, \textbf{Kimia Path24}, for image classification and retrieval in digital pathology. We use the whole scan images of 24 different tissue textures to generate 1,325 test patches of size 1000$\times$1000 (0.5mm$\times$0.5mm). Training data can be generated according to preferences of algorithm designer and can range from approximately 27,000 to over 50,000 patches if the preset parameters are adopted. We propose a compound patch-and-scan accuracy measurement that makes achieving high accuracies quite challenging. In addition, we set the benchmarking line by applying LBP, dictionary approach and convolutional neural nets (CNNs) and report their results. The highest accuracy was 41.80\% for CNN.
Retrieving Similar X-Ray Images from Big Image Data Using Radon Barcodes with Single Projections
Jan 02, 2017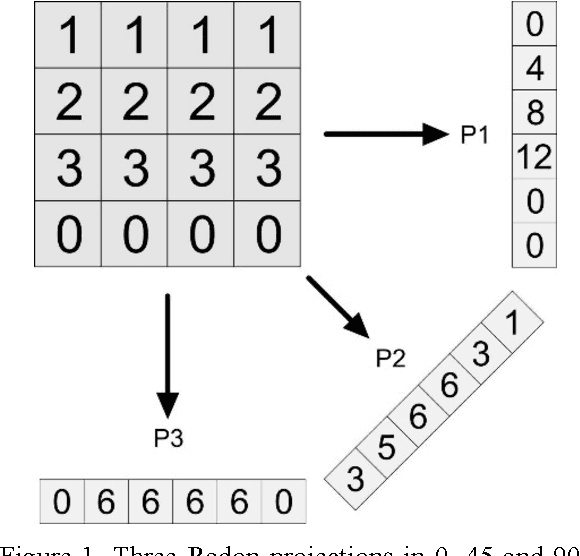
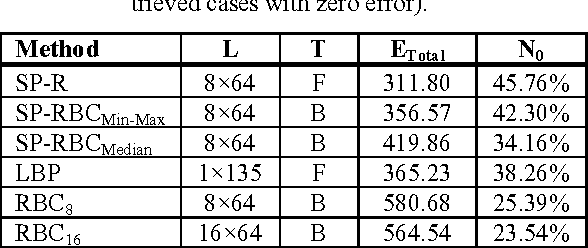

Abstract:The idea of Radon barcodes (RBC) has been introduced recently. In this paper, we propose a content-based image retrieval approach for big datasets based on Radon barcodes. Our method (Single Projection Radon Barcode, or SP-RBC) uses only a few Radon single projections for each image as global features that can serve as a basis for weak learners. This is our most important contribution in this work, which improves the results of the RBC considerably. As a matter of fact, only one projection of an image, as short as a single SURF feature vector, can already achieve acceptable results. Nevertheless, using multiple projections in a long vector will not deliver anticipated improvements. To exploit the information inherent in each projection, our method uses the outcome of each projection separately and then applies more precise local search on the small subset of retrieved images. We have tested our method using IRMA 2009 dataset a with 14,400 x-ray images as part of imageCLEF initiative. Our approach leads to a substantial decrease in the error rate in comparison with other non-learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge