Guillaume Theaud
Clinical-ComBAT: a diffusion-weighted MRI harmonization method for clinical applications
Nov 06, 2025Abstract:Diffusion-weighted magnetic resonance imaging (DW-MRI) derived scalar maps are effective for assessing neurodegenerative diseases and microstructural properties of white matter in large number of brain conditions. However, DW-MRI inherently limits the combination of data from multiple acquisition sites without harmonization to mitigate scanner-specific biases. While the widely used ComBAT method reduces site effects in research, its reliance on linear covariate relationships, homogeneous populations, fixed site numbers, and well populated sites constrains its clinical use. To overcome these limitations, we propose Clinical-ComBAT, a method designed for real-world clinical scenarios. Clinical-ComBAT harmonizes each site independently, enabling flexibility as new data and clinics are introduced. It incorporates a non-linear polynomial data model, site-specific harmonization referenced to a normative site, and variance priors adaptable to small cohorts. It further includes hyperparameter tuning and a goodness-of-fit metric for harmonization assessment. We demonstrate its effectiveness on simulated and real data, showing improved alignment of diffusion metrics and enhanced applicability for normative modeling.
ComBAT Harmonization for diffusion MRI: Challenges and Best Practices
May 19, 2025Abstract:Over the years, ComBAT has become the standard method for harmonizing MRI-derived measurements, with its ability to compensate for site-related additive and multiplicative biases while preserving biological variability. However, ComBAT relies on a set of assumptions that, when violated, can result in flawed harmonization. In this paper, we thoroughly review ComBAT's mathematical foundation, outlining these assumptions, and exploring their implications for the demographic composition necessary for optimal results. Through a series of experiments involving a slightly modified version of ComBAT called Pairwise-ComBAT tailored for normative modeling applications, we assess the impact of various population characteristics, including population size, age distribution, the absence of certain covariates, and the magnitude of additive and multiplicative factors. Based on these experiments, we present five essential recommendations that should be carefully considered to enhance consistency and supporting reproducibility, two essential factors for open science, collaborative research, and real-life clinical deployment.
Tractography filtering using autoencoders
Oct 07, 2020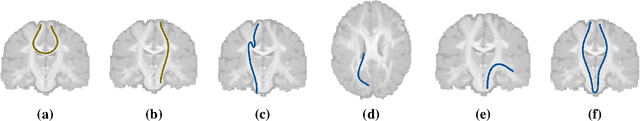
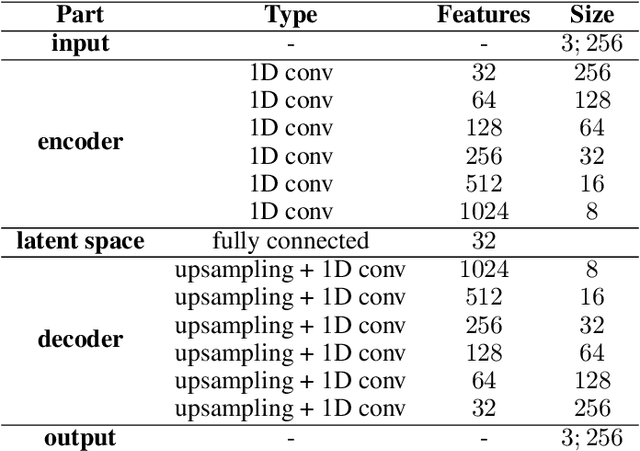
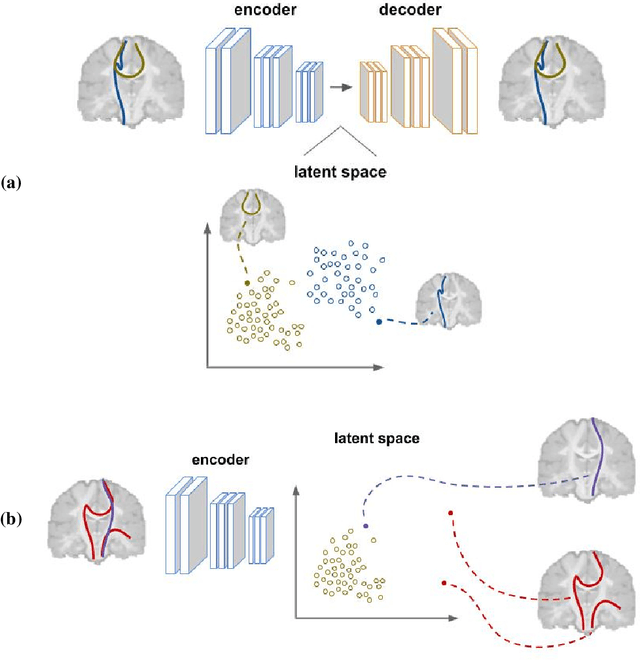
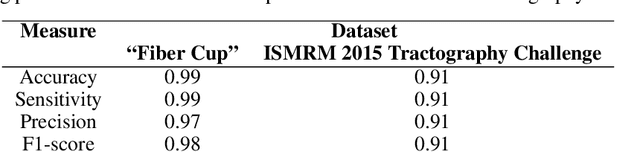
Abstract:Current brain white matter fiber tracking techniques show a number of problems, including: generating large proportions of streamlines that do not accurately describe the underlying anatomy; extracting streamlines that are not supported by the underlying diffusion signal; and under-representing some fiber populations, among others. In this paper, we describe a novel unsupervised learning method to filter streamlines from diffusion MRI tractography, and hence, to obtain more reliable tractograms. We show that a convolutional neural network autoencoder provides a straightforward and elegant way to learn a robust representation of brain streamlines, which can be used to filter undesired samples with a nearest neighbor algorithm. Our method, dubbed FINTA (Filtering in Tractography using Autoencoders) comes with several key advantages: training does not need labeled data, as it uses raw tractograms, it is fast and easily reproducible, it does not rely on the input diffusion MRI data, and thus, does not suffer from domain adaptation issues. We demonstrate the ability of FINTA to discriminate between "plausible" and "implausible" streamlines as well as to recover individual streamline group instances from a raw tractogram, from both synthetic and real human brain diffusion MRI tractography data, including partial tractograms. Results reveal that FINTA has a superior filtering performance compared to state-of-the-art methods. Together, this work brings forward a new deep learning framework in tractography based on autoencoders, and shows how it can be applied for filtering purposes. It sets the foundations for opening up new prospects towards more accurate and robust tractometry and connectivity diffusion MRI analyses, which may ultimately lead to improve the imaging of the white matter anatomy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge