Folkert W. Asselbergs
Interpretable phenotyping of Heart Failure patients with Dutch discharge letters
May 30, 2025Abstract:Objective: Heart failure (HF) patients present with diverse phenotypes affecting treatment and prognosis. This study evaluates models for phenotyping HF patients based on left ventricular ejection fraction (LVEF) classes, using structured and unstructured data, assessing performance and interpretability. Materials and Methods: The study analyzes all HF hospitalizations at both Amsterdam UMC hospitals (AMC and VUmc) from 2015 to 2023 (33,105 hospitalizations, 16,334 patients). Data from AMC were used for model training, and from VUmc for external validation. The dataset was unlabelled and included tabular clinical measurements and discharge letters. Silver labels for LVEF classes were generated by combining diagnosis codes, echocardiography results, and textual mentions. Gold labels were manually annotated for 300 patients for testing. Multiple Transformer-based (black-box) and Aug-Linear (white-box) models were trained and compared with baselines on structured and unstructured data. To evaluate interpretability, two clinicians annotated 20 discharge letters by highlighting information they considered relevant for LVEF classification. These were compared to SHAP and LIME explanations from black-box models and the inherent explanations of Aug-Linear models. Results: BERT-based and Aug-Linear models, using discharge letters alone, achieved the highest classification results (AUC=0.84 for BERT, 0.81 for Aug-Linear on external validation), outperforming baselines. Aug-Linear explanations aligned more closely with clinicians' explanations than post-hoc explanations on black-box models. Conclusions: Discharge letters emerged as the most informative source for phenotyping HF patients. Aug-Linear models matched black-box performance while providing clinician-aligned interpretability, supporting their use in transparent clinical decision-making.
Multimodal Learning for Cardiovascular Risk Prediction using EHR Data
Aug 27, 2020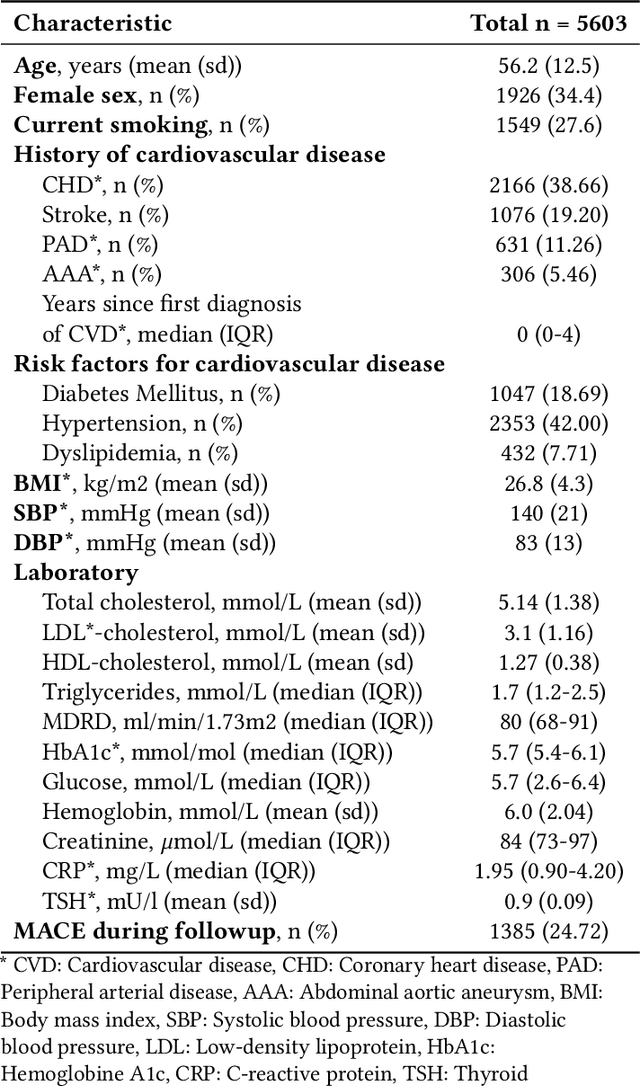
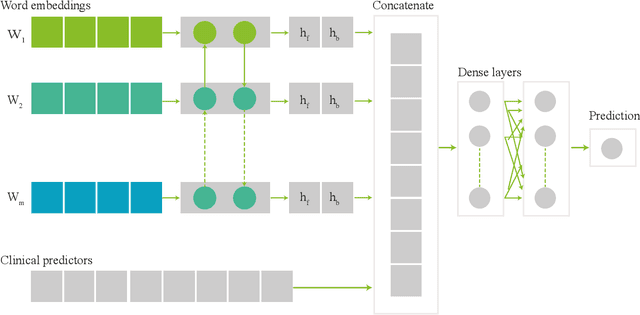
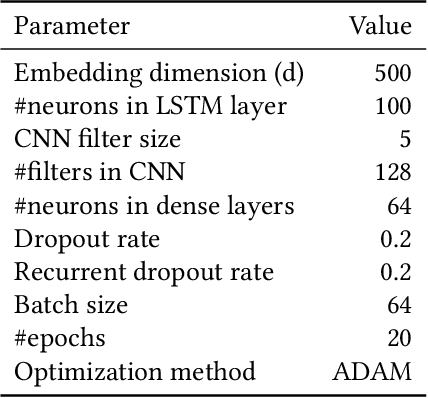
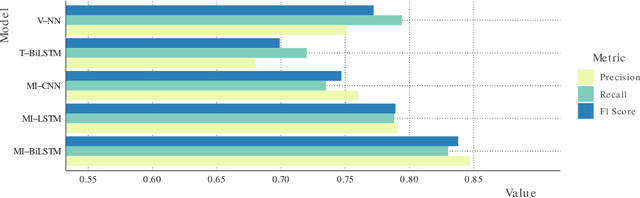
Abstract:Electronic health records (EHRs) contain structured and unstructured data of significant clinical and research value. Various machine learning approaches have been developed to employ information in EHRs for risk prediction. The majority of these attempts, however, focus on structured EHR fields and lose the vast amount of information in the unstructured texts. To exploit the potential information captured in EHRs, in this study we propose a multimodal recurrent neural network model for cardiovascular risk prediction that integrates both medical texts and structured clinical information. The proposed multimodal bidirectional long short-term memory (BiLSTM) model concatenates word embeddings to classical clinical predictors before applying them to a final fully connected neural network. In the experiments, we compare performance of different deep neural network (DNN) architectures including convolutional neural network and long short-term memory in scenarios of using clinical variables and chest X-ray radiology reports. Evaluated on a data set of real world patients with manifest vascular disease or at high-risk for cardiovascular disease, the proposed BiLSTM model demonstrates state-of-the-art performance and outperforms other DNN baseline architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge