Federico Ariel
ChronoRoot 2.0: An Open AI-Powered Platform for 2D Temporal Plant Phenotyping
Apr 20, 2025



Abstract:The analysis of plant developmental plasticity, including root system architecture, is fundamental to understanding plant adaptability and development, particularly in the context of climate change and agricultural sustainability. While significant advances have been made in plant phenotyping technologies, comprehensive temporal analysis of root development remains challenging, with most existing solutions providing either limited throughput or restricted structural analysis capabilities. Here, we present ChronoRoot 2.0, an integrated open-source platform that combines affordable hardware with advanced artificial intelligence to enable sophisticated temporal plant phenotyping. The system introduces several major advances, offering an integral perspective of seedling development: (i) simultaneous multi-organ tracking of six distinct plant structures, (ii) quality control through real-time validation, (iii) comprehensive architectural measurements including novel gravitropic response parameters, and (iv) dual specialized user interfaces for both architectural analysis and high-throughput screening. We demonstrate the system's capabilities through three use cases for Arabidopsis thaliana: characterization of circadian growth patterns under different light conditions, detailed analysis of gravitropic responses in transgenic plants, and high-throughput screening of etiolation responses across multiple genotypes. ChronoRoot 2.0 maintains its predecessor's advantages of low cost and modularity while significantly expanding its capabilities, making sophisticated temporal phenotyping more accessible to the broader plant science community. The system's open-source nature, combined with extensive documentation and containerized deployment options, ensures reproducibility and enables community-driven development of new analytical capabilities.
Arabidopsis roots segmentation based on morphological operations and CRFs
Apr 25, 2017
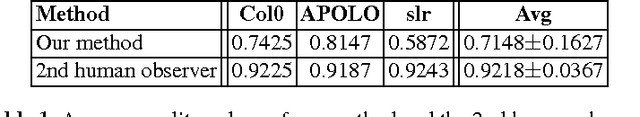
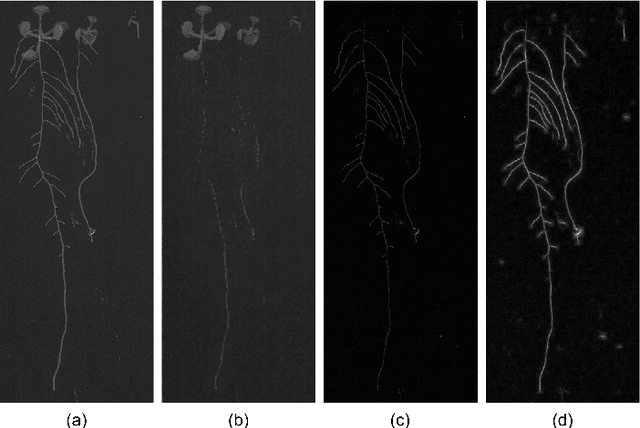

Abstract:Arabidopsis thaliana is a plant species widely utilized by scientists to estimate the impact of genetic differences in root morphological features. For this purpose, images of this plant after genetic modifications are taken to study differences in the root architecture. This task requires manual segmentations of radicular structures, although this is a particularly tedious and time-consuming labor. In this work, we present an unsupervised method for Arabidopsis thaliana root segmentation based on morphological operations and fully-connected Conditional Random Fields. Although other approaches have been proposed to this purpose, all of them are based on more complex and expensive imaging modalities. Our results prove that our method can be easily applied over images taken using conventional scanners, with a minor user intervention. A first data set, our results and a fully open source implementation are available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge