Farzad Khalvati
Localized Perturbations For Weakly-Supervised Segmentation of Glioma Brain Tumours
Nov 29, 2021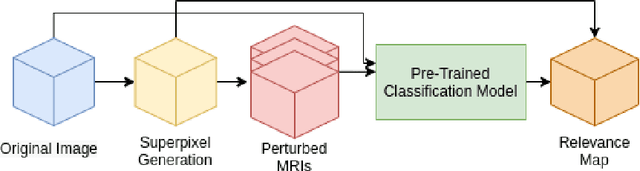
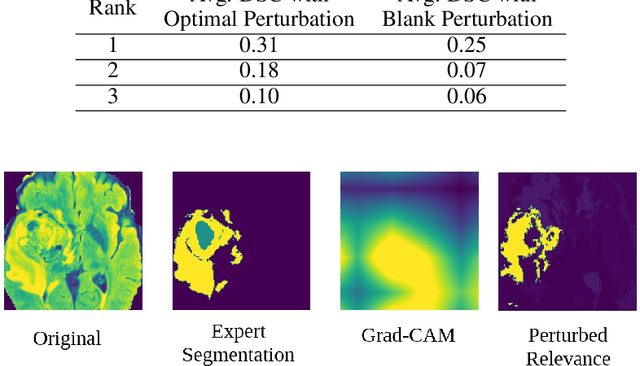
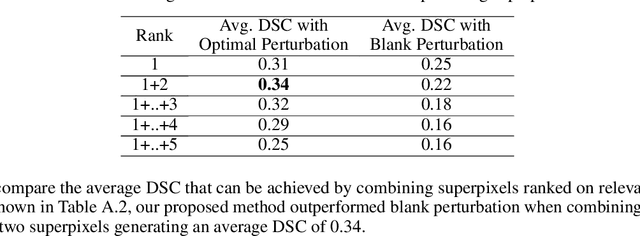

Abstract:Deep convolutional neural networks (CNNs) have become an essential tool in the medical imaging-based computer-aided diagnostic pipeline. However, training accurate and reliable CNNs requires large fine-grain annotated datasets. To alleviate this, weakly-supervised methods can be used to obtain local information from global labels. This work proposes the use of localized perturbations as a weakly-supervised solution to extract segmentation masks of brain tumours from a pretrained 3D classification model. Furthermore, we propose a novel optimal perturbation method that exploits 3D superpixels to find the most relevant area for a given classification using a U-net architecture. Our method achieved a Dice similarity coefficient (DSC) of 0.44 when compared with expert annotations. When compared against Grad-CAM, our method outperformed both in visualization and localization ability of the tumour region, with Grad-CAM only achieving 0.11 average DSC.
Vanishing Twin GAN: How training a weak Generative Adversarial Network can improve semi-supervised image classification
Mar 03, 2021
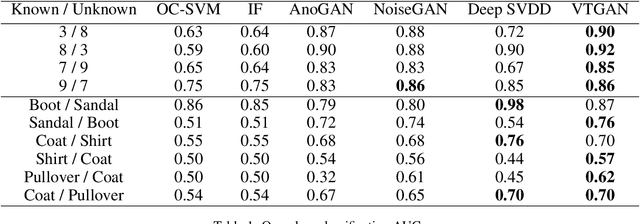

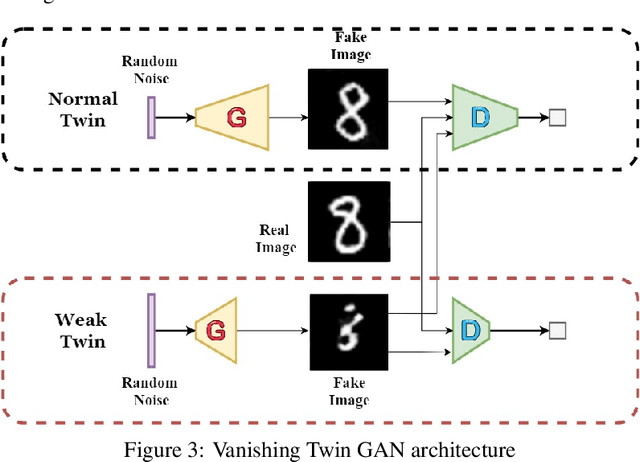
Abstract:Generative Adversarial Networks can learn the mapping of random noise to realistic images in a semi-supervised framework. This mapping ability can be used for semi-supervised image classification to detect images of an unknown class where there is no training data to be used for supervised classification. However, if the unknown class shares similar characteristics to the known class(es), GANs can learn to generalize and generate images that look like both classes. This generalization ability can hinder the classification performance. In this work, we propose the Vanishing Twin GAN. By training a weak GAN and using its generated output image parallel to the regular GAN, the Vanishing Twin training improves semi-supervised image classification where image similarity can hurt classification tasks.
Multi-class Generative Adversarial Nets for Semi-supervised Image Classification
Feb 22, 2021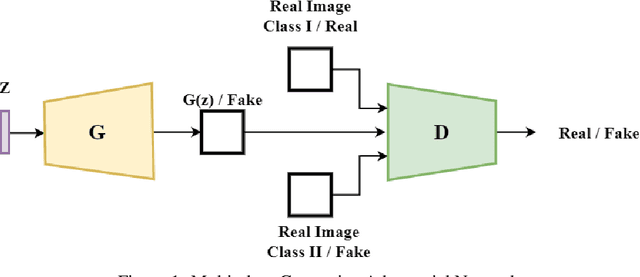
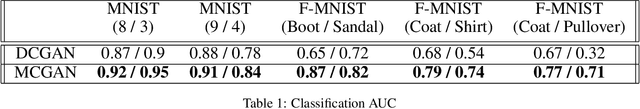
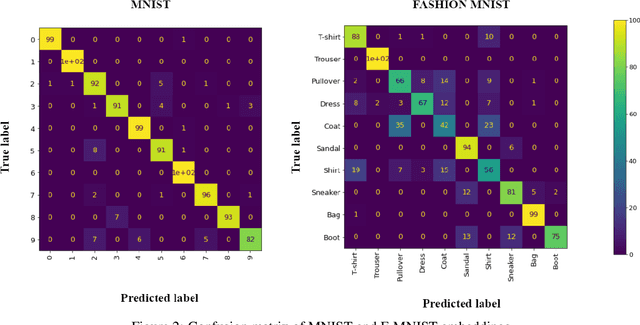
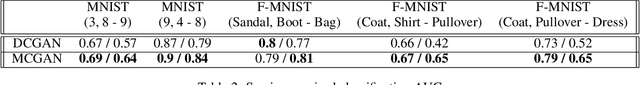
Abstract:From generating never-before-seen images to domain adaptation, applications of Generative Adversarial Networks (GANs) spread wide in the domain of vision and graphics problems. With the remarkable ability of GANs in learning the distribution and generating images of a particular class, they can be used for semi-supervised classification tasks. However, the problem is that if two classes of images share similar characteristics, the GAN might learn to generalize and hinder the classification of the two classes. In this paper, we use various images from MNIST and Fashion-MNIST datasets to illustrate how similar images cause the GAN to generalize, leading to the poor classification of images. We propose a modification to the traditional training of GANs that allows for improved multi-class classification in similar classes of images in a semi-supervised learning framework.
A Transfer Learning Based Active Learning Framework for Brain Tumor Classification
Nov 16, 2020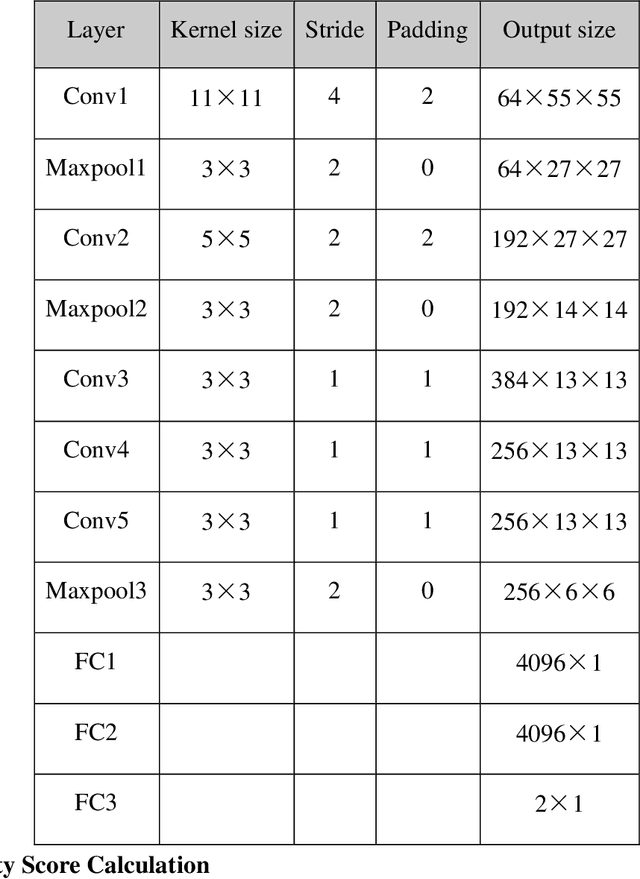
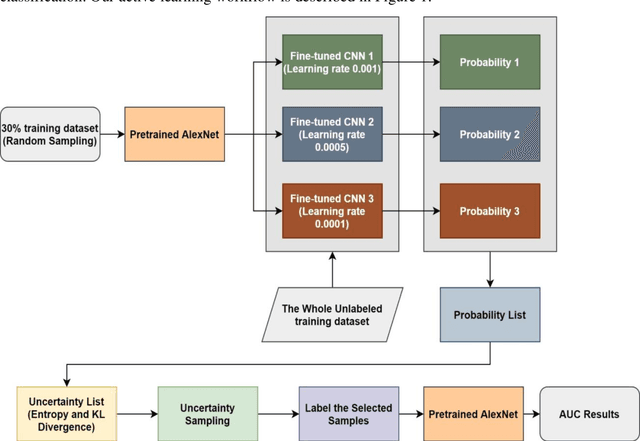
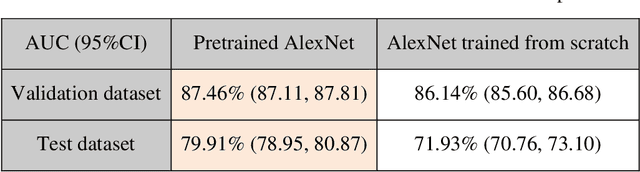
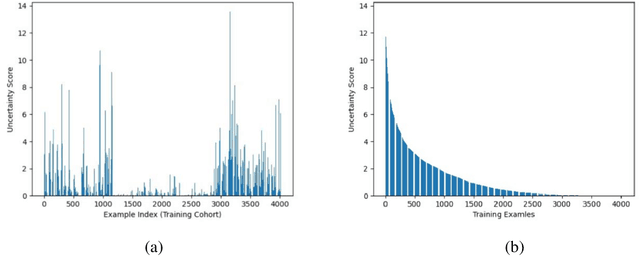
Abstract:Brain tumor is one of the leading causes of cancer-related death globally among children and adults. Precise classification of brain tumor grade (low-grade and high-grade glioma) at early stage plays a key role in successful prognosis and treatment planning. With recent advances in deep learning, Artificial Intelligence-enabled brain tumor grading systems can assist radiologists in the interpretation of medical images within seconds. The performance of deep learning techniques is, however, highly depended on the size of the annotated dataset. It is extremely challenging to label a large quantity of medical images given the complexity and volume of medical data. In this work, we propose a novel transfer learning based active learning framework to reduce the annotation cost while maintaining stability and robustness of the model performance for brain tumor classification. We employed a 2D slice-based approach to train and finetune our model on the Magnetic Resonance Imaging (MRI) training dataset of 203 patients and a validation dataset of 66 patients which was used as the baseline. With our proposed method, the model achieved Area Under Receiver Operating Characteristic (ROC) Curve (AUC) of 82.89% on a separate test dataset of 66 patients, which was 2.92% higher than the baseline AUC while saving at least 40% of labeling cost. In order to further examine the robustness of our method, we created a balanced dataset, which underwent the same procedure. The model achieved AUC of 82% compared with AUC of 78.48% for the baseline, which reassures the robustness and stability of our proposed transfer learning augmented with active learning framework while significantly reducing the size of training data.
RANDGAN: Randomized Generative Adversarial Network for Detection of COVID-19 in Chest X-ray
Oct 06, 2020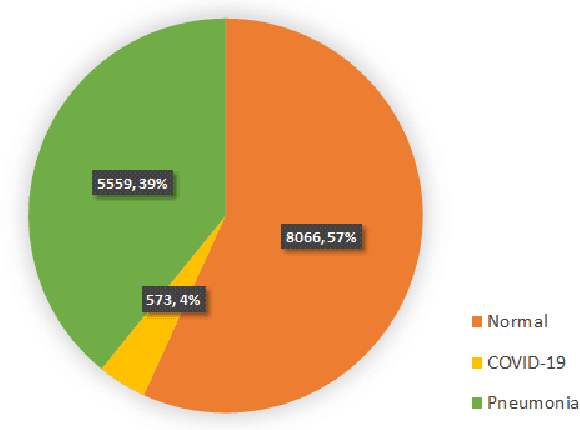
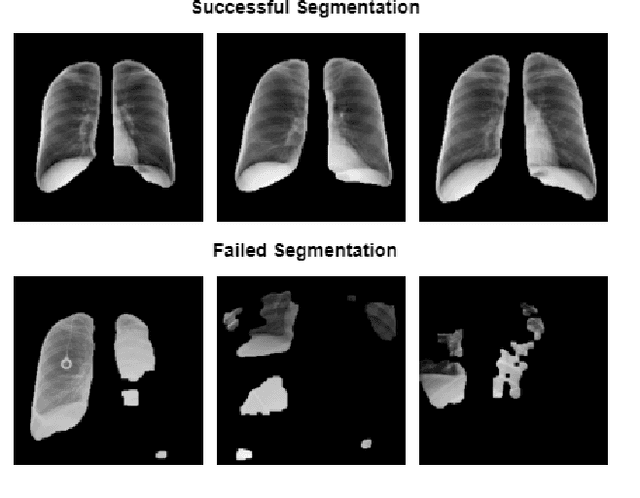
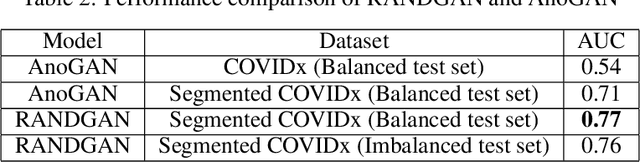
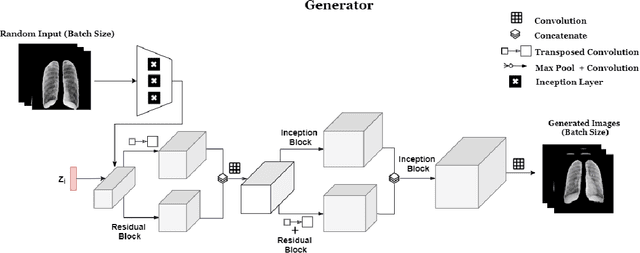
Abstract:COVID-19 spread across the globe at an immense rate has left healthcare systems incapacitated to diagnose and test patients at the needed rate. Studies have shown promising results for detection of COVID-19 from viral bacterial pneumonia in chest X-rays. Automation of COVID-19 testing using medical images can speed up the testing process of patients where health care systems lack sufficient numbers of the reverse-transcription polymerase chain reaction (RT-PCR) tests. Supervised deep learning models such as convolutional neural networks (CNN) need enough labeled data for all classes to correctly learn the task of detection. Gathering labeled data is a cumbersome task and requires time and resources which could further strain health care systems and radiologists at the early stages of a pandemic such as COVID-19. In this study, we propose a randomized generative adversarial network (RANDGAN) that detects images of an unknown class (COVID-19) from known and labelled classes (Normal and Viral Pneumonia) without the need for labels and training data from the unknown class of images (COVID-19). We used the largest publicly available COVID-19 chest X-ray dataset, COVIDx, which is comprised of Normal, Pneumonia, and COVID-19 images from multiple public databases. In this work, we use transfer learning to segment the lungs in the COVIDx dataset. Next, we show why segmentation of the region of interest (lungs) is vital to correctly learn the task of classification, specifically in datasets that contain images from different resources as it is the case for the COVIDx dataset. Finally, we show improved results in detection of COVID-19 cases using our generative model (RANDGAN) compared to conventional generative adversarial networks (GANs) for anomaly detection in medical images, improving the area under the ROC curve from 0.71 to 0.77.
Evaluating Knowledge Transfer in Neural Network for Medical Images
Sep 17, 2020
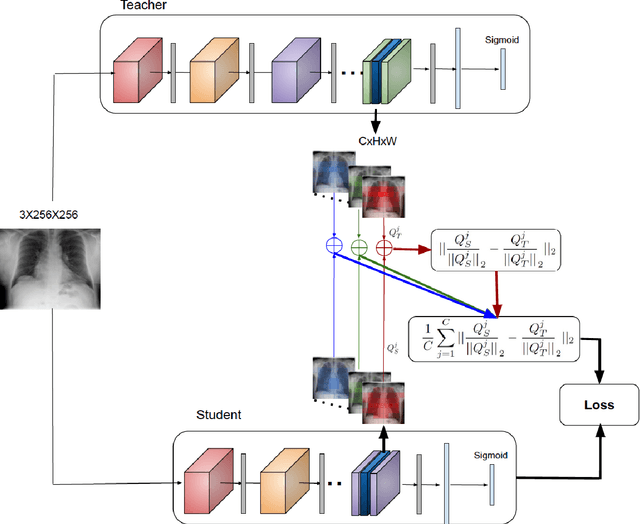
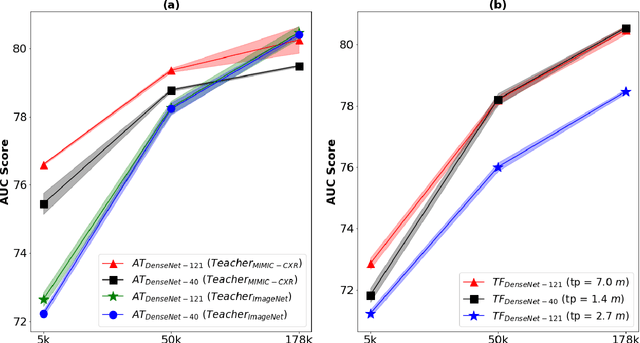
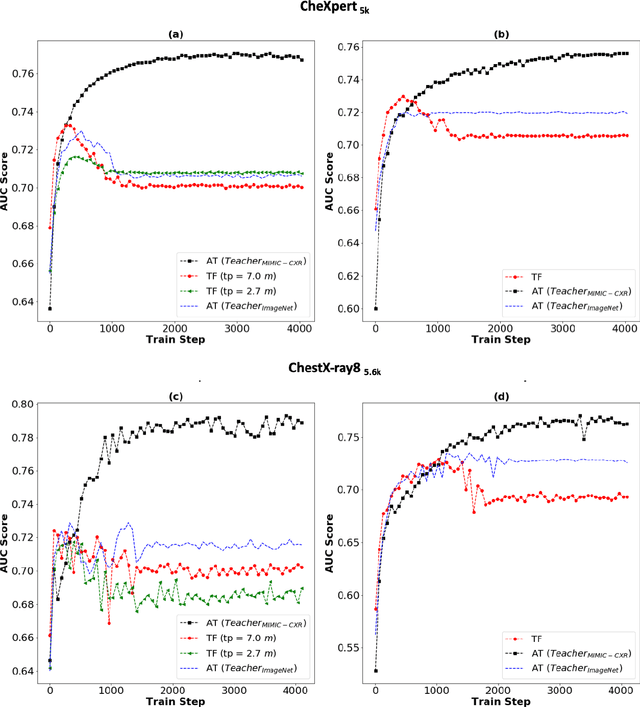
Abstract:Deep learning and knowledge transfer techniques have permeated the field of medical imaging and are considered as key approaches for revolutionizing diagnostic imaging practices. However, there are still challenges for the successful integration of deep learning into medical imaging tasks due to a lack of large annotated imaging data. To address this issue, we propose a teacher-student learning framework to transfer knowledge from a carefully pre-trained convolutional neural network (CNN) teacher to a student CNN. In this study, we explore the performance of knowledge transfer in the medical imaging setting. We investigate the proposed network's performance when the student network is trained on a small dataset (target dataset) as well as when teacher's and student's domains are distinct. The performances of the CNN models are evaluated on three medical imaging datasets including Diabetic Retinopathy, CheXpert, and ChestX-ray8. Our results indicate that the teacher-student learning framework outperforms transfer learning for small imaging datasets. Particularly, the teacher-student learning framework improves the area under the ROC Curve (AUC) of the CNN model on a small sample of CheXpert (n=5k) by 4% and on ChestX-ray8 (n=5.6k) by 9%. In addition to small training data size, we also demonstrate a clear advantage of the teacher-student learning framework in the medical imaging setting compared to transfer learning. We observe that the teacher-student network holds a great promise not only to improve the performance of diagnosis but also to reduce overfitting when the dataset is small.
A Brief Review of Deep Multi-task Learning and Auxiliary Task Learning
Jul 02, 2020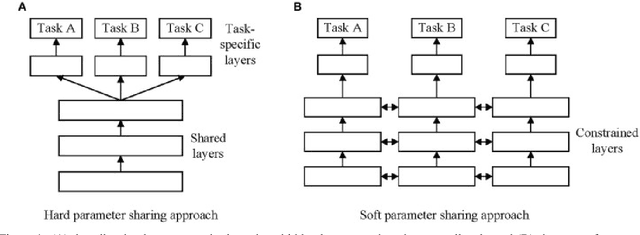
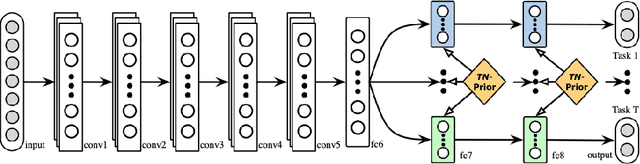
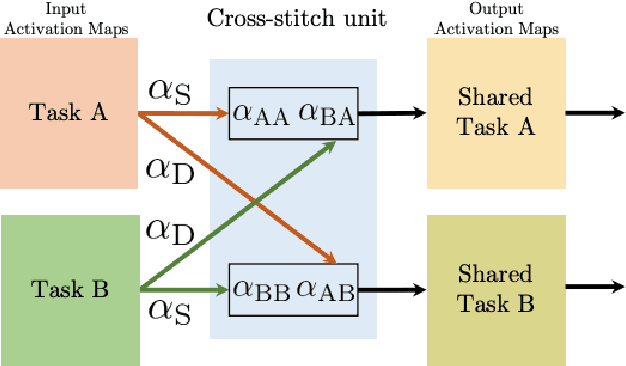
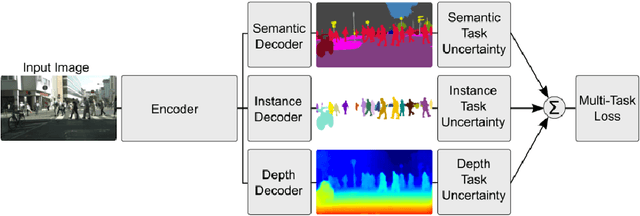
Abstract:Multi-task learning (MTL) optimizes several learning tasks simultaneously and leverages their shared information to improve generalization and the prediction of the model for each task. Auxiliary tasks can be added to the main task to ultimately boost the performance. In this paper, we provide a brief review on the recent deep multi-task learning (dMTL) approaches followed by methods on selecting useful auxiliary tasks that can be used in dMTL to improve the performance of the model for the main task.
A Modified AUC for Training Convolutional Neural Networks: Taking Confidence into Account
Jun 08, 2020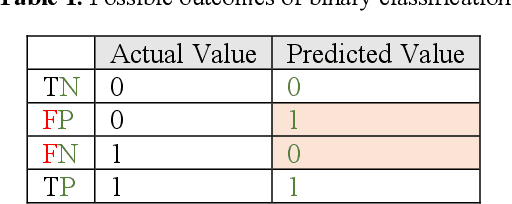
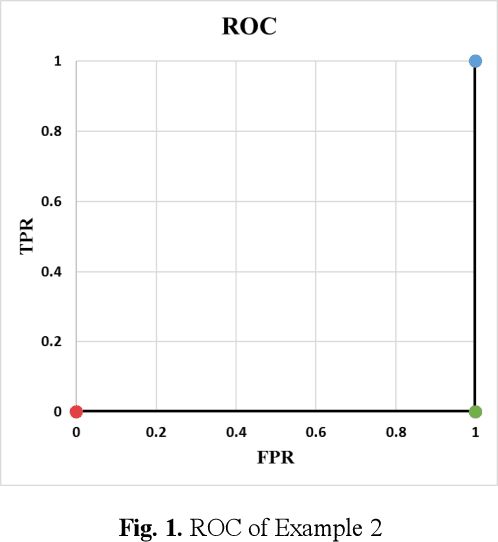
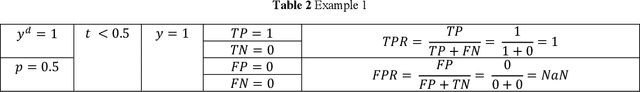
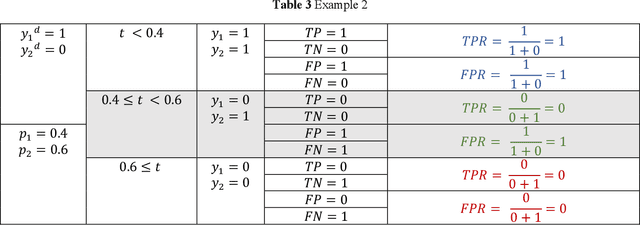
Abstract:Receiver operating characteristic (ROC) curve is an informative tool in binary classification and Area Under ROC Curve (AUC) is a popular metric for reporting performance of binary classifiers. In this paper, first we present a comprehensive review of ROC curve and AUC metric. Next, we propose a modified version of AUC that takes confidence of the model into account and at the same time, incorporates AUC into Binary Cross Entropy (BCE) loss used for training a Convolutional neural Network for classification tasks. We demonstrate this on two datasets: MNIST and prostate MRI. Furthermore, we have published GenuineAI, a new python library, which provides the functions for conventional AUC and the proposed modified AUC along with metrics including sensitivity, specificity, recall, precision, and F1 for each point of the ROC curve.
Inception Augmentation Generative Adversarial Network
Jun 05, 2020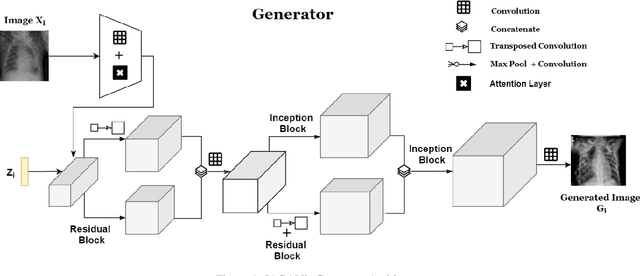
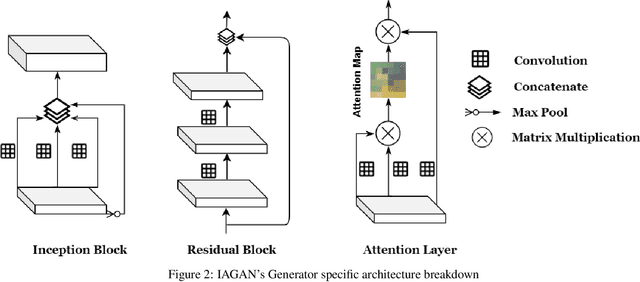
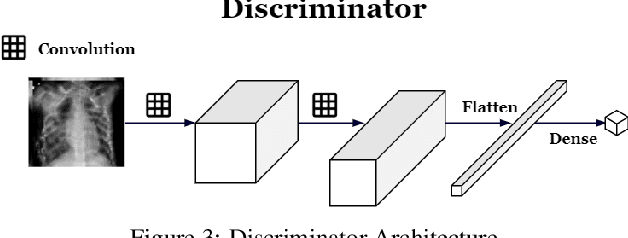
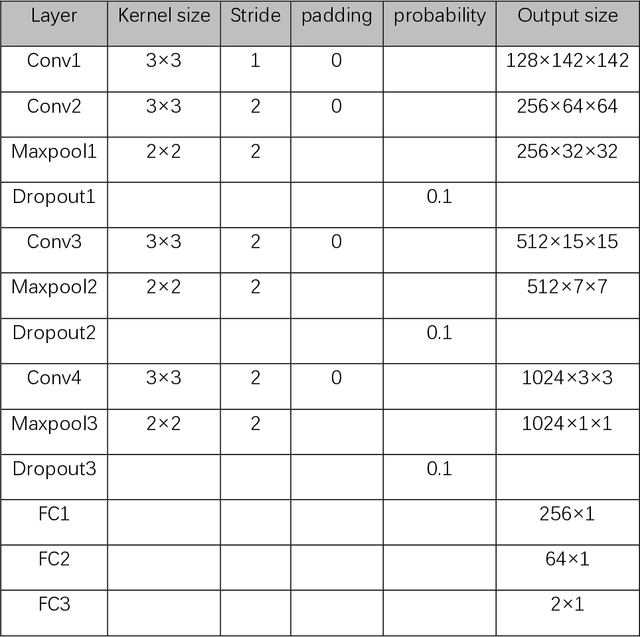
Abstract:Successful training of convolutional neural networks (CNNs) requires a substantial amount of data. With small datasets, networks generalize poorly. Data Augmentation techniques improve the generalizability of neural networks by using existing training data more effectively. Standard data augmentation methods, however, produce limited plausible alternative data. Generative Adversarial Networks (GANs) have been utilized to generate new data and improve CNN performance. Nevertheless, generative models have not been used for augmenting data to improve the training of another generative model. In this work, we propose a new GAN architecture for semi-supervised augmentation of chest X-rays for the detection of pneumonia. We show that the proposed GAN can augment data for a specific class of images (pneumonia) using images from both classes (pneumonia and normal) in an image domain (chest X-rays). We demonstrate that using our proposed GAN-based data augmentation method significantly improves the performance of the state-of-the-art anomaly detection architecture, AnoGAN, in detecting pneumonia in chest X-rays, increasing AUC from 0.83 to 0.88.
A Comprehensive Study of Data Augmentation Strategies for Prostate Cancer Detection in Diffusion-weighted MRI using Convolutional Neural Networks
Jun 01, 2020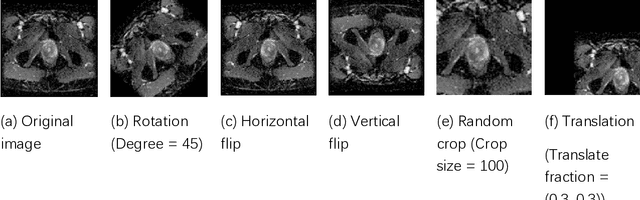
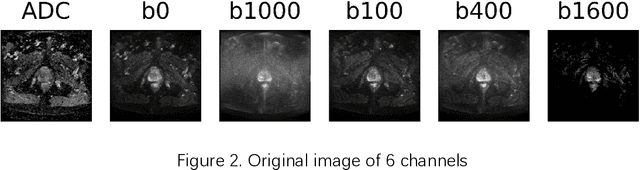
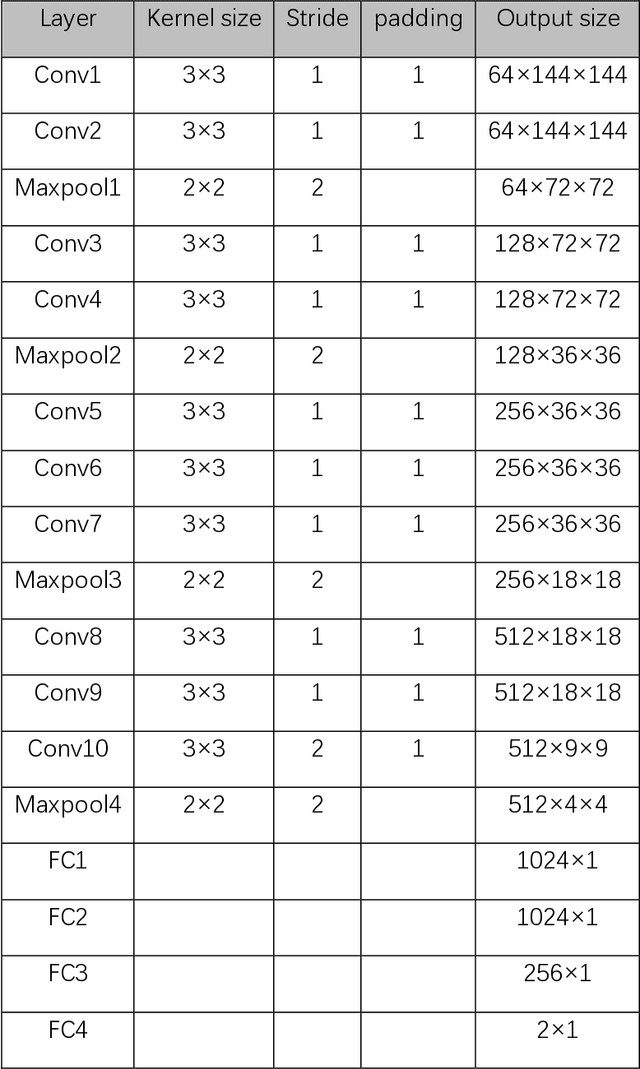
Abstract:Data augmentation refers to a group of techniques whose goal is to battle limited amount of available data to improve model generalization and push sample distribution toward the true distribution. While different augmentation strategies and their combinations have been investigated for various computer vision tasks in the context of deep learning, a specific work in the domain of medical imaging is rare and to the best of our knowledge, there has been no dedicated work on exploring the effects of various augmentation methods on the performance of deep learning models in prostate cancer detection. In this work, we have statically applied five most frequently used augmentation techniques (random rotation, horizontal flip, vertical flip, random crop, and translation) to prostate Diffusion-weighted Magnetic Resonance Imaging training dataset of 217 patients separately and evaluated the effect of each method on the accuracy of prostate cancer detection. The augmentation algorithms were applied independently to each data channel and a shallow as well as a deep Convolutional Neural Network (CNN) were trained on the five augmented sets separately. We used Area Under Receiver Operating Characteristic (ROC) curve (AUC) to evaluate the performance of the trained CNNs on a separate test set of 95 patients, using a validation set of 102 patients for finetuning. The shallow network outperformed the deep network with the best 2D slice-based AUC of 0.85 obtained by the rotation method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge