Farzad Abdolhosseini
CVNets: High Performance Library for Computer Vision
Jun 04, 2022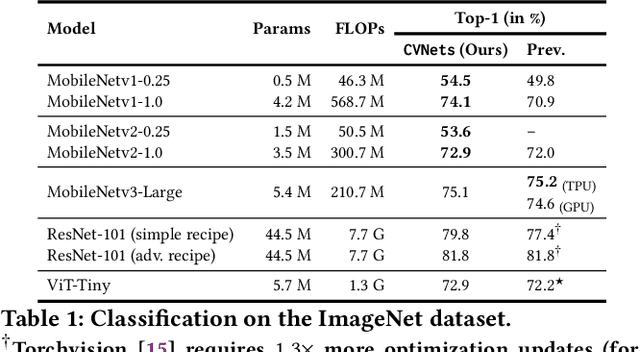
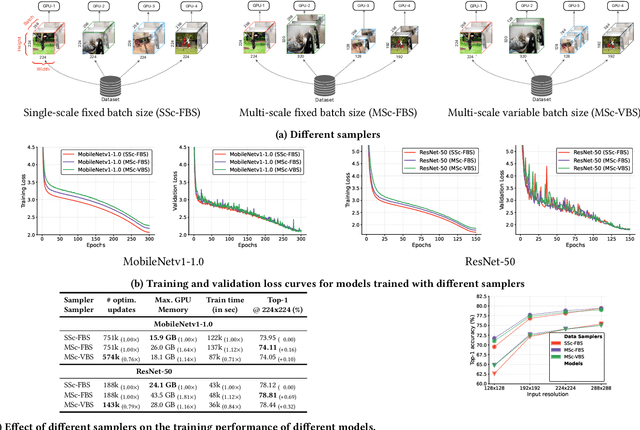
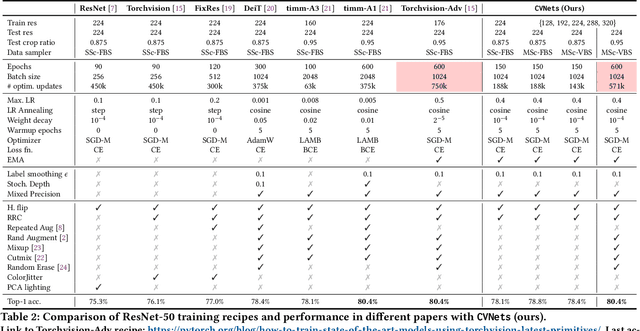
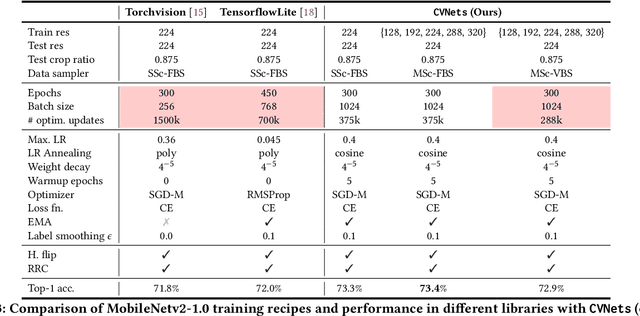
Abstract:We introduce CVNets, a high-performance open-source library for training deep neural networks for visual recognition tasks, including classification, detection, and segmentation. CVNets supports image and video understanding tools, including data loading, data transformations, novel data sampling methods, and implementations of several standard networks with similar or better performance than previous studies. Our source code is available at: \url{https://github.com/apple/ml-cvnets}.
Cell Identity Codes: Understanding Cell Identity from Gene Expression Profiles using Deep Neural Networks
Jun 13, 2018Abstract:Understanding cell identity is an important task in many biomedical areas. Expression patterns of specific marker genes have been used to characterize some limited cell types, but exclusive markers are not available for many cell types. A second approach is to use machine learning to discriminate cell types based on the whole gene expression profiles (GEPs). The accuracies of simple classification algorithms such as linear discriminators or support vector machines are limited due to the complexity of biological systems. We used deep neural networks to analyze 1040 GEPs from 16 different human tissues and cell types. After comparing different architectures, we identified a specific structure of deep autoencoders that can encode a GEP into a vector of 30 numeric values, which we call the cell identity code (CIC). The original GEP can be reproduced from the CIC with an accuracy comparable to technical replicates of the same experiment. Although we use an unsupervised approach to train the autoencoder, we show different values of the CIC are connected to different biological aspects of the cell, such as different pathways or biological processes. This network can use CIC to reproduce the GEP of the cell types it has never seen during the training. It also can resist some noise in the measurement of the GEP. Furthermore, we introduce classifier autoencoder, an architecture that can accurately identify cell type based on the GEP or the CIC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge