Erik Smistad
Lightweight image segmentation for echocardiography
Sep 03, 2025



Abstract:Accurate segmentation of the left ventricle in echocardiography can enable fully automatic extraction of clinical measurements such as volumes and ejection fraction. While models configured by nnU-Net perform well, they are large and slow, thus limiting real-time use. We identified the most effective components of nnU-Net for cardiac segmentation through an ablation study, incrementally evaluating data augmentation schemes, architectural modifications, loss functions, and post-processing techniques. Our analysis revealed that simple affine augmentations and deep supervision drive performance, while complex augmentations and large model capacity offer diminishing returns. Based on these insights, we developed a lightweight U-Net (2M vs 33M parameters) that achieves statistically equivalent performance to nnU-Net on CAMUS (N=500) with Dice scores of 0.93/0.85/0.89 vs 0.93/0.86/0.89 for LV/MYO/LA ($p>0.05$), while being 16 times smaller and 4 times faster (1.35ms vs 5.40ms per frame) than the default nnU-Net configuration. Cross-dataset evaluation on an internal dataset (N=311) confirms comparable generalization.
Generative augmentations for improved cardiac ultrasound segmentation using diffusion models
Feb 27, 2025Abstract:One of the main challenges in current research on segmentation in cardiac ultrasound is the lack of large and varied labeled datasets and the differences in annotation conventions between datasets. This makes it difficult to design robust segmentation models that generalize well to external datasets. This work utilizes diffusion models to create generative augmentations that can significantly improve diversity of the dataset and thus the generalisability of segmentation models without the need for more annotated data. The augmentations are applied in addition to regular augmentations. A visual test survey showed that experts cannot clearly distinguish between real and fully generated images. Using the proposed generative augmentations, segmentation robustness was increased when training on an internal dataset and testing on an external dataset with an improvement of over 20 millimeters in Hausdorff distance. Additionally, the limits of agreement for automatic ejection fraction estimation improved by up to 20% of absolute ejection fraction value on out of distribution cases. These improvements come exclusively from the increased variation of the training data using the generative augmentations, without modifying the underlying machine learning model. The augmentation tool is available as an open source Python library at https://github.com/GillesVanDeVyver/EchoGAINS.
Regional quality estimation for echocardiography using deep learning
Aug 01, 2024Abstract:Automatic estimation of cardiac ultrasound image quality can be beneficial for guiding operators and ensuring the accuracy of clinical measurements. Previous work often fails to distinguish the view correctness of the echocardiogram from the image quality. Additionally, previous studies only provide a global image quality value, which limits their practical utility. In this work, we developed and compared three methods to estimate image quality: 1) classic pixel-based metrics like the generalized contrast-to-noise ratio (gCNR) on myocardial segments as region of interest and left ventricle lumen as background, obtained using a U-Net segmentation 2) local image coherence derived from a U-Net model that predicts coherence from B-Mode images 3) a deep convolutional network that predicts the quality of each region directly in an end-to-end fashion. We evaluate each method against manual regional image quality annotations by three experienced cardiologists. The results indicate poor performance of the gCNR metric, with Spearman correlation to the annotations of \r{ho} = 0.24. The end-to-end learning model obtains the best result, \r{ho} = 0.69, comparable to the inter-observer correlation, \r{ho} = 0.63. Finally, the coherence-based method, with \r{ho} = 0.58, outperformed the classical metrics and is more generic than the end-to-end approach.
Segmentation of Non-Small Cell Lung Carcinomas: Introducing DRU-Net and Multi-Lens Distortion
Jun 20, 2024



Abstract:Considering the increased workload in pathology laboratories today, automated tools such as artificial intelligence models can help pathologists with their tasks and ease the workload. In this paper, we are proposing a segmentation model (DRU-Net) that can provide a delineation of human non-small cell lung carcinomas and an augmentation method that can improve classification results. The proposed model is a fused combination of truncated pre-trained DenseNet201 and ResNet101V2 as a patch-wise classifier followed by a lightweight U-Net as a refinement model. We have used two datasets (Norwegian Lung Cancer Biobank and Haukeland University Hospital lung cancer cohort) to create our proposed model. The DRU-Net model achieves an average of 0.91 Dice similarity coefficient. The proposed spatial augmentation method (multi-lens distortion) improved the network performance by 3%. Our findings show that choosing image patches that specifically include regions of interest leads to better results for the patch-wise classifier compared to other sampling methods. The qualitative analysis showed that the DRU-Net model is generally successful in detecting the tumor. On the test set, some of the cases showed areas of false positive and false negative segmentation in the periphery, particularly in tumors with inflammatory and reactive changes.
Immunohistochemistry guided segmentation of benign epithelial cells, in situ lesions, and invasive epithelial cells in breast cancer slides
Nov 22, 2023Abstract:Digital pathology enables automatic analysis of histopathological sections using artificial intelligence (AI). Automatic evaluation could improve diagnostic efficiency and help find associations between morphological features and clinical outcome. For development of such prediction models, identifying invasive epithelial cells, and separating these from benign epithelial cells and in situ lesions would be the first step. In this study, we aimed to develop an AI model for segmentation of epithelial cells in sections from breast cancer. We generated epithelial ground truth masks by restaining hematoxylin and eosin (HE) sections with cytokeratin (CK) AE1/AE3, and by pathologists' annotations. HE/CK image pairs were used to train a convolutional neural network, and data augmentation was used to make the model more robust. Tissue microarrays (TMAs) from 839 patients, and whole slide images from two patients were used for training and evaluation of the models. The sections were derived from four cohorts of breast cancer patients. TMAs from 21 patients from a fifth cohort was used as a second test set. In quantitative evaluation, a mean Dice score of 0.70, 0.79, and 0.75 for invasive epithelial cells, benign epithelial cells, and in situ lesions, respectively, were achieved. In qualitative scoring (0-5) by pathologists, results were best for all epithelium and invasive epithelium, with scores of 4.7 and 4.4. Scores for benign epithelium and in situ lesions were 3.7 and 2.0. The proposed model segmented epithelial cells in HE stained breast cancer slides well, but further work is needed for accurate division between the classes. Immunohistochemistry, together with pathologists' annotations, enabled the creation of accurate ground truths. The model is made freely available in FastPathology and the code is available at https://github.com/AICAN-Research/breast-epithelium-segmentation
Towards Robust Cardiac Segmentation using Graph Convolutional Networks
Oct 02, 2023



Abstract:Fully automatic cardiac segmentation can be a fast and reproducible method to extract clinical measurements from an echocardiography examination. The U-Net architecture is the current state-of-the-art deep learning architecture for medical segmentation and can segment cardiac structures in real-time with average errors comparable to inter-observer variability. However, this architecture still generates large outliers that are often anatomically incorrect. This work uses the concept of graph convolutional neural networks that predict the contour points of the structures of interest instead of labeling each pixel. We propose a graph architecture that uses two convolutional rings based on cardiac anatomy and show that this eliminates anatomical incorrect multi-structure segmentations on the publicly available CAMUS dataset. Additionally, this work contributes with an ablation study on the graph convolutional architecture and an evaluation of clinical measurements on the clinical HUNT4 dataset. Finally, we propose to use the inter-model agreement of the U-Net and the graph network as a predictor of both the input and segmentation quality. We show this predictor can detect out-of-distribution and unsuitable input images in real-time. Source code is available online: https://github.com/gillesvntnu/GCN_multistructure
Hybrid guiding: A multi-resolution refinement approach for semantic segmentation of gigapixel histopathological images
Dec 07, 2021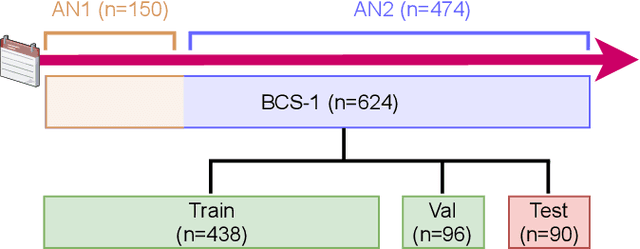
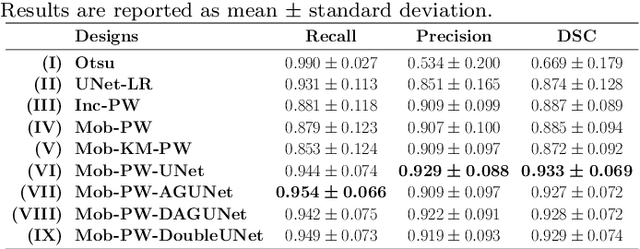
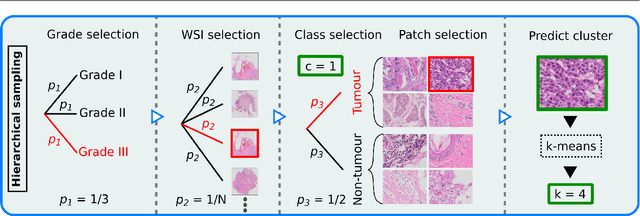
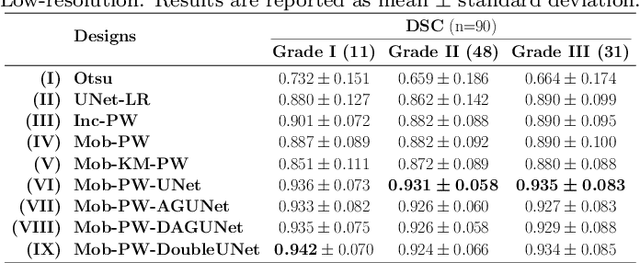
Abstract:Histopathological cancer diagnostics has become more complex, and the increasing number of biopsies is a challenge for most pathology laboratories. Thus, development of automatic methods for evaluation of histopathological cancer sections would be of value. In this study, we used 624 whole slide images (WSIs) of breast cancer from a Norwegian cohort. We propose a cascaded convolutional neural network design, called H2G-Net, for semantic segmentation of gigapixel histopathological images. The design involves a detection stage using a patch-wise method, and a refinement stage using a convolutional autoencoder. To validate the design, we conducted an ablation study to assess the impact of selected components in the pipeline on tumour segmentation. Guiding segmentation, using hierarchical sampling and deep heatmap refinement, proved to be beneficial when segmenting the histopathological images. We found a significant improvement when using a refinement network for postprocessing the generated tumour segmentation heatmaps. The overall best design achieved a Dice score of 0.933 on an independent test set of 90 WSIs. The design outperformed single-resolution approaches, such as cluster-guided, patch-wise high-resolution classification using MobileNetV2 (0.872) and a low-resolution U-Net (0.874). In addition, segmentation on a representative x400 WSI took ~58 seconds, using only the CPU. The findings demonstrate the potential of utilizing a refinement network to improve patch-wise predictions. The solution is efficient and does not require overlapping patch inference or ensembling. Furthermore, we showed that deep neural networks can be trained using a random sampling scheme that balances on multiple different labels simultaneously, without the need of storing patches on disk. Future work should involve more efficient patch generation and sampling, as well as improved clustering.
Code-free development and deployment of deep segmentation models for digital pathology
Nov 16, 2021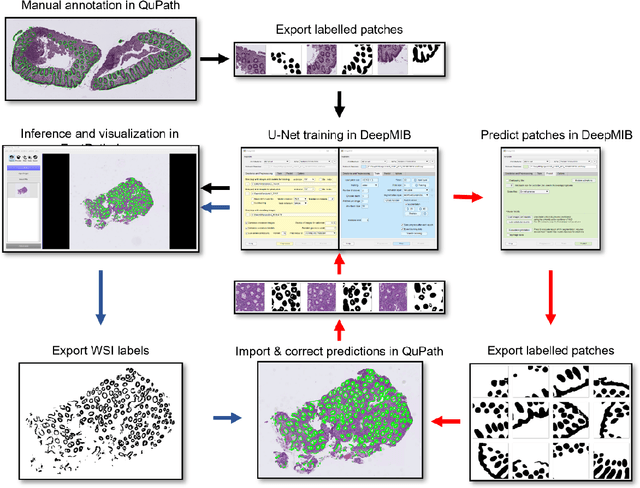
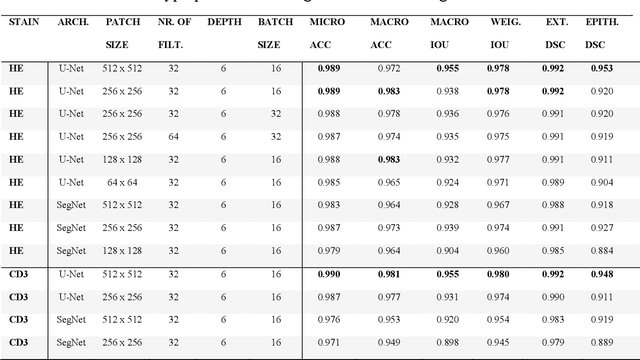
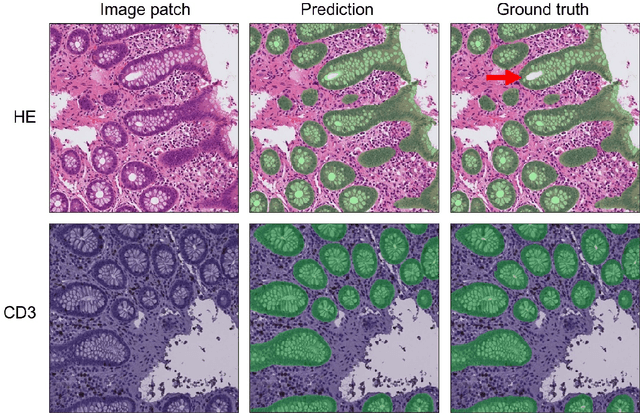
Abstract:Application of deep learning on histopathological whole slide images (WSIs) holds promise of improving diagnostic efficiency and reproducibility but is largely dependent on the ability to write computer code or purchase commercial solutions. We present a code-free pipeline utilizing free-to-use, open-source software (QuPath, DeepMIB, and FastPathology) for creating and deploying deep learning-based segmentation models for computational pathology. We demonstrate the pipeline on a use case of separating epithelium from stroma in colonic mucosa. A dataset of 251 annotated WSIs, comprising 140 hematoxylin-eosin (HE)-stained and 111 CD3 immunostained colon biopsy WSIs, were developed through active learning using the pipeline. On a hold-out test set of 36 HE and 21 CD3-stained WSIs a mean intersection over union score of 96.6% and 95.3% was achieved on epithelium segmentation. We demonstrate pathologist-level segmentation accuracy and clinical acceptable runtime performance and show that pathologists without programming experience can create near state-of-the-art segmentation solutions for histopathological WSIs using only free-to-use software. The study further demonstrates the strength of open-source solutions in its ability to create generalizable, open pipelines, of which trained models and predictions can seamlessly be exported in open formats and thereby used in external solutions. All scripts, trained models, a video tutorial, and the full dataset of 251 WSIs with ~31k epithelium annotations are made openly available at https://github.com/andreped/NoCodeSeg to accelerate research in the field.
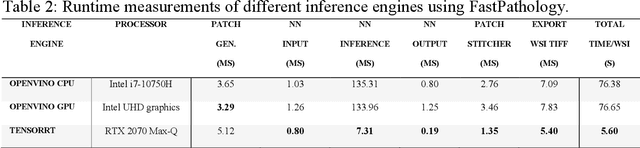
FastPathology: An open-source platform for deep learning-based research and decision support in digital pathology
Nov 11, 2020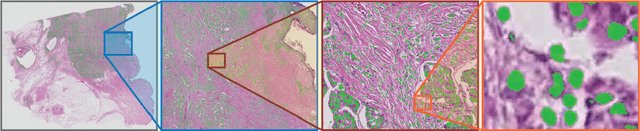
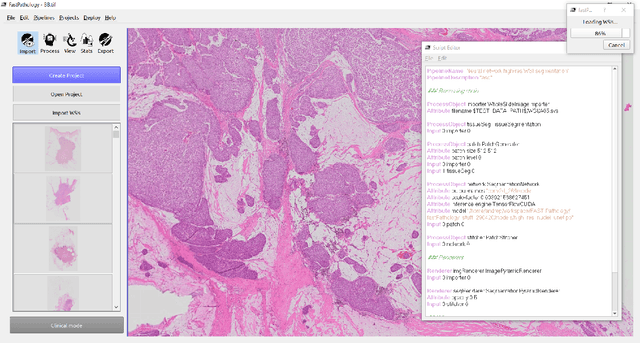
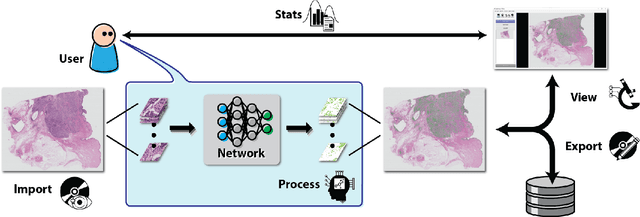
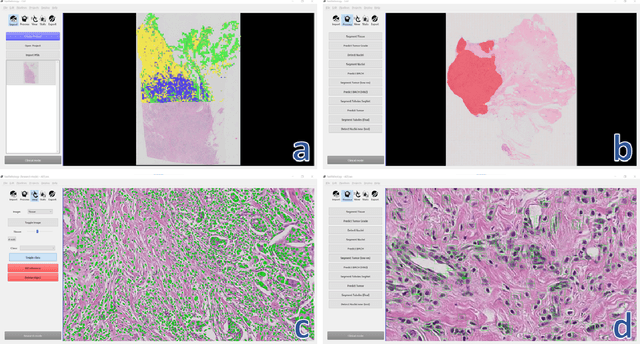
Abstract:Deep convolutional neural networks (CNNs) are the current state-of-the-art for digital analysis of histopathological images. The large size of whole-slide microscopy images (WSIs) requires advanced memory handling to read, display and process these images. There are several open-source platforms for working with WSIs, but few support deployment of CNN models. These applications use third-party solutions for inference, making them less user-friendly and unsuitable for high-performance image analysis. To make deployment of CNNs user-friendly and feasible on low-end machines, we have developed a new platform, FastPathology, using the FAST framework and C++. It minimizes memory usage for reading and processing WSIs, deployment of CNN models, and real-time interactive visualization of results. Runtime experiments were conducted on four different use cases, using different architectures, inference engines, hardware configurations and operating systems. Memory usage for reading, visualizing, zooming and panning a WSI were measured, using FastPathology and three existing platforms. FastPathology performed similarly in terms of memory to the other C++ based application, while using considerably less than the two Java-based platforms. The choice of neural network model, inference engine, hardware and processors influenced runtime considerably. Thus, FastPathology includes all steps needed for efficient visualization and processing of WSIs in a single application, including inference of CNNs with real-time display of the results. Source code, binary releases and test data can be found online on GitHub at https://github.com/SINTEFMedtek/FAST-Pathology/.
LU-Net: a multi-task network to improve the robustness of segmentation of left ventriclular structures by deep learning in 2D echocardiography
Apr 04, 2020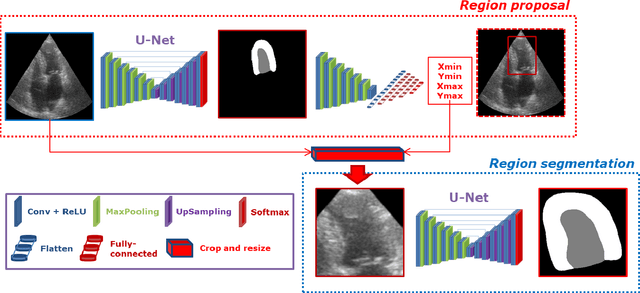
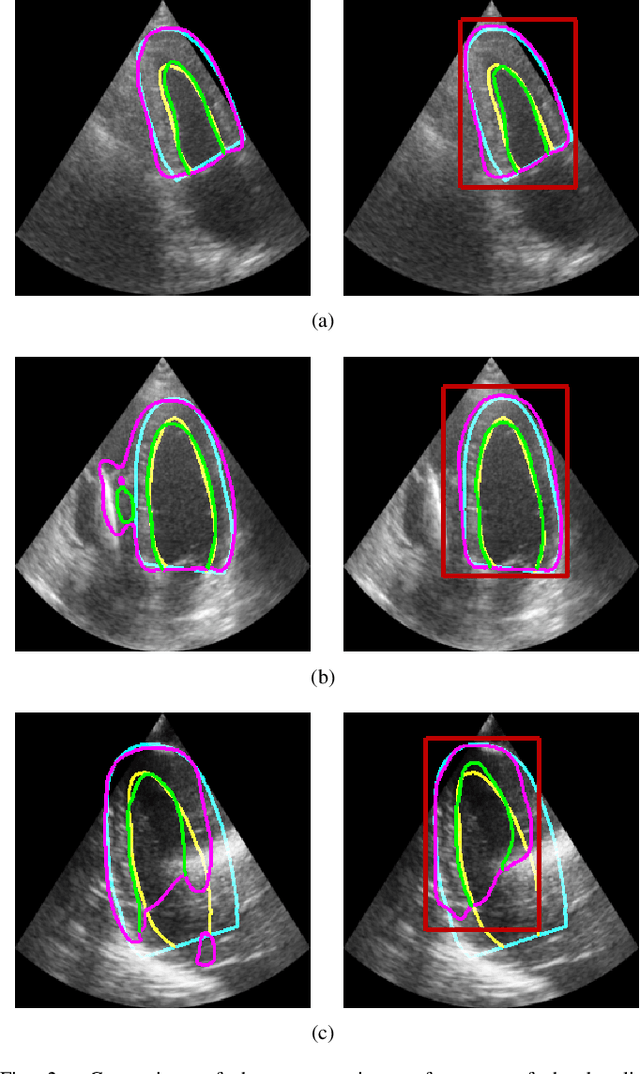
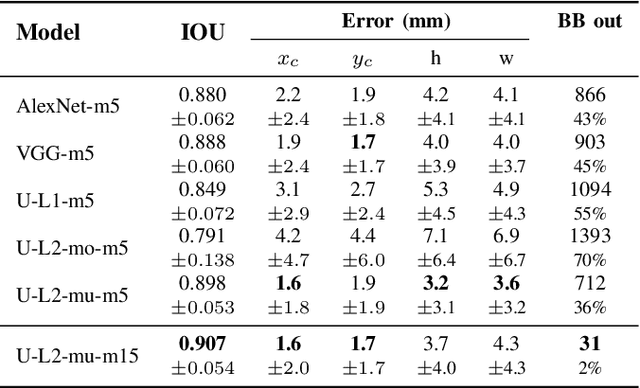
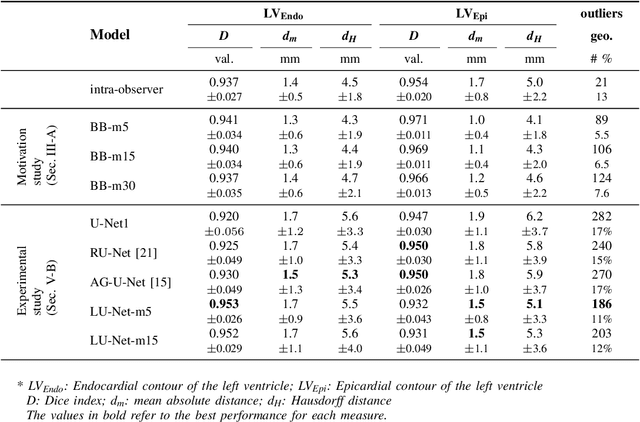
Abstract:Segmentation of cardiac structures is one of the fundamental steps to estimate volumetric indices of the heart. This step is still performed semi-automatically in clinical routine, and is thus prone to inter- and intra-observer variability. Recent studies have shown that deep learning has the potential to perform fully automatic segmentation. However, the current best solutions still suffer from a lack of robustness. In this work, we introduce an end-to-end multi-task network designed to improve the overall accuracy of cardiac segmentation while enhancing the estimation of clinical indices and reducing the number of outliers. Results obtained on a large open access dataset show that our method outperforms the current best performing deep learning solution and achieved an overall segmentation accuracy lower than the intra-observer variability for the epicardial border (i.e. on average a mean absolute error of 1.5mm and a Hausdorff distance of 5.1mm) with 11% of outliers. Moreover, we demonstrate that our method can closely reproduce the expert analysis for the end-diastolic and end-systolic left ventricular volumes, with a mean correlation of 0.96 and a mean absolute error of 7.6ml. Concerning the ejection fraction of the left ventricle, results are more contrasted with a mean correlation coefficient of 0.83 and an absolute mean error of 5.0%, producing scores that are slightly below the intra-observer margin. Based on this observation, areas for improvement are suggested.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge