Ekansh Chauhan
Contrasting Low and High-Resolution Features for HER2 Scoring using Deep Learning
Mar 28, 2025Abstract:Breast cancer, the most common malignancy among women, requires precise detection and classification for effective treatment. Immunohistochemistry (IHC) biomarkers like HER2, ER, and PR are critical for identifying breast cancer subtypes. However, traditional IHC classification relies on pathologists' expertise, making it labor-intensive and subject to significant inter-observer variability. To address these challenges, this study introduces the India Pathology Breast Cancer Dataset (IPD-Breast), comprising of 1,272 IHC slides (HER2, ER, and PR) aimed at automating receptor status classification. The primary focus is on developing predictive models for HER2 3-way classification (0, Low, High) to enhance prognosis. Evaluation of multiple deep learning models revealed that an end-to-end ConvNeXt network utilizing low-resolution IHC images achieved an AUC, F1, and accuracy of 91.79%, 83.52%, and 83.56%, respectively, for 3-way classification, outperforming patch-based methods by over 5.35% in F1 score. This study highlights the potential of simple yet effective deep learning techniques to significantly improve accuracy and reproducibility in breast cancer classification, supporting their integration into clinical workflows for better patient outcomes.
Multiple Instance Learning for Glioma Diagnosis using Hematoxylin and Eosin Whole Slide Images: An Indian Cohort Study
Mar 08, 2024



Abstract:The effective management of brain tumors relies on precise typing, subtyping, and grading. This study advances patient care with findings from rigorous multiple instance learning experimentations across various feature extractors and aggregators in brain tumor histopathology. It establishes new performance benchmarks in glioma subtype classification across multiple datasets, including a novel dataset focused on the Indian demographic (IPD- Brain), providing a valuable resource for existing research. Using a ResNet-50, pretrained on histopathology datasets for feature extraction, combined with the Double-Tier Feature Distillation (DTFD) feature aggregator, our approach achieves state-of-the-art AUCs of 88.08 on IPD-Brain and 95.81 on the TCGA-Brain dataset, respectively, for three-way glioma subtype classification. Moreover, it establishes new benchmarks in grading and detecting IHC molecular biomarkers (IDH1R132H, TP53, ATRX, Ki-67) through H&E stained whole slide images for the IPD-Brain dataset. The work also highlights a significant correlation between the model decision-making processes and the diagnostic reasoning of pathologists, underscoring its capability to mimic professional diagnostic procedures.
LRH-Net: A Multi-Level Knowledge Distillation Approach for Low-Resource Heart Network
Apr 11, 2022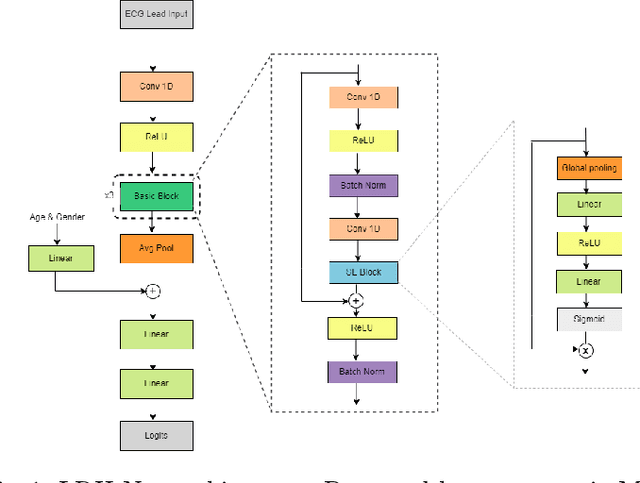
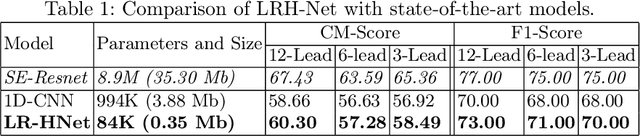
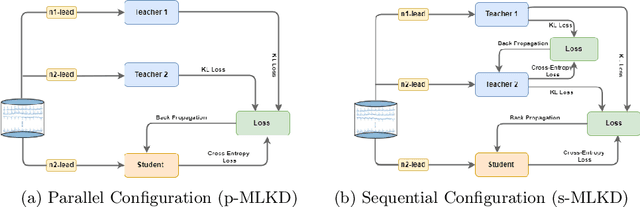
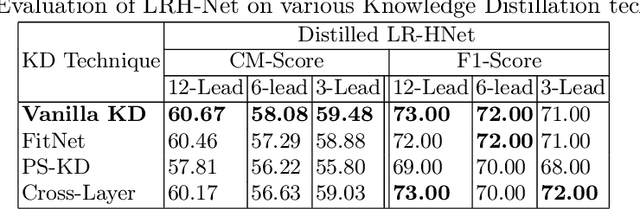
Abstract:An electrocardiogram (ECG) monitors the electrical activity generated by the heart and is used to detect fatal cardiovascular diseases (CVDs). Conventionally, to capture the precise electrical activity, clinical experts use multiple-lead ECGs (typically 12 leads). But in recent times, large-size deep learning models have been used to detect these diseases. However, such models require heavy compute resources like huge memory and long inference time. To alleviate these shortcomings, we propose a low-parameter model, named Low Resource Heart-Network (LRH-Net), which uses fewer leads to detect ECG anomalies in a resource-constrained environment. A multi-level knowledge distillation process is used on top of that to get better generalization performance on our proposed model. The multi-level knowledge distillation process distills the knowledge to LRH-Net trained on a reduced number of leads from higher parameter (teacher) models trained on multiple leads to reduce the performance gap. The proposed model is evaluated on the PhysioNet-2020 challenge dataset with constrained input. The parameters of the LRH-Net are 106x less than our teacher model for detecting CVDs. The performance of the LRH-Net was scaled up to 3.2% and the inference time scaled down by 75% compared to the teacher model. In contrast to the compute- and parameter-intensive deep learning techniques, the proposed methodology uses a subset of ECG leads using the low resource LRH-Net, making it eminently suitable for deployment on edge devices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge