Dmitri B. Chklovskii
A Similarity-preserving Neural Network Trained on Transformed Images Recapitulates Salient Features of the Fly Motion Detection Circuit
Feb 10, 2021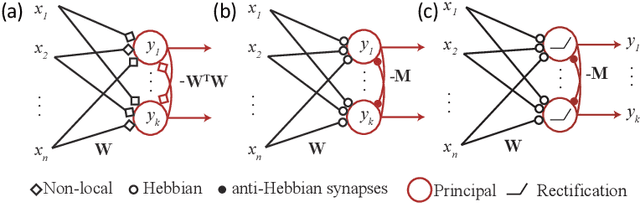
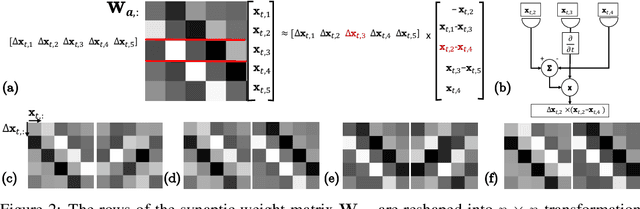
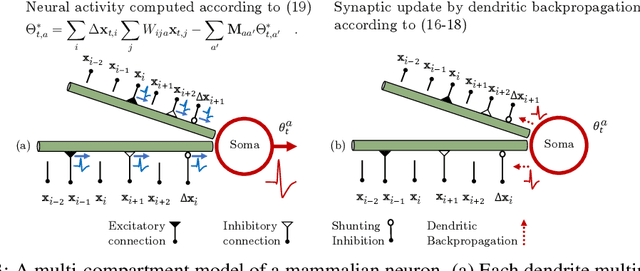
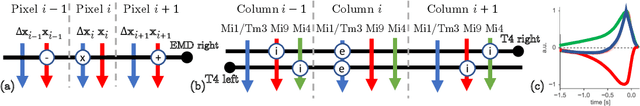
Abstract:Learning to detect content-independent transformations from data is one of the central problems in biological and artificial intelligence. An example of such problem is unsupervised learning of a visual motion detector from pairs of consecutive video frames. Rao and Ruderman formulated this problem in terms of learning infinitesimal transformation operators (Lie group generators) via minimizing image reconstruction error. Unfortunately, it is difficult to map their model onto a biologically plausible neural network (NN) with local learning rules. Here we propose a biologically plausible model of motion detection. We also adopt the transformation-operator approach but, instead of reconstruction-error minimization, start with a similarity-preserving objective function. An online algorithm that optimizes such an objective function naturally maps onto an NN with biologically plausible learning rules. The trained NN recapitulates major features of the well-studied motion detector in the fly. In particular, it is consistent with the experimental observation that local motion detectors combine information from at least three adjacent pixels, something that contradicts the celebrated Hassenstein-Reichardt model.
A Neural Network with Local Learning Rules for Minor Subspace Analysis
Feb 10, 2021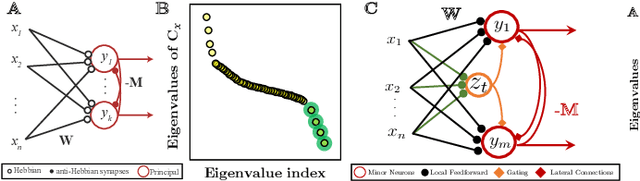
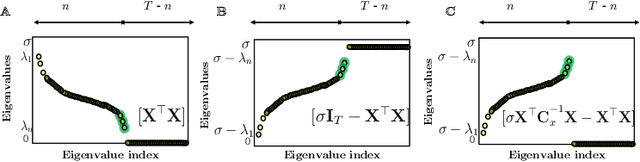
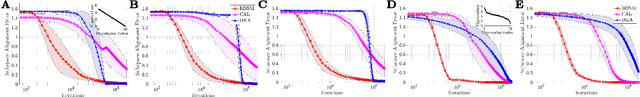
Abstract:The development of neuromorphic hardware and modeling of biological neural networks requires algorithms with local learning rules. Artificial neural networks using local learning rules to perform principal subspace analysis (PSA) and clustering have recently been derived from principled objective functions. However, no biologically plausible networks exist for minor subspace analysis (MSA), a fundamental signal processing task. MSA extracts the lowest-variance subspace of the input signal covariance matrix. Here, we introduce a novel similarity matching objective for extracting the minor subspace, Minor Subspace Similarity Matching (MSSM). Moreover, we derive an adaptive MSSM algorithm that naturally maps onto a novel neural network with local learning rules and gives numerical results showing that our method converges at a competitive rate.
A biologically plausible neural network for local supervision in cortical microcircuits
Nov 30, 2020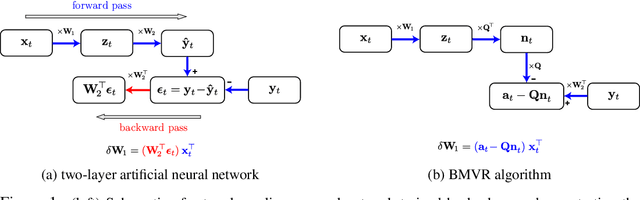
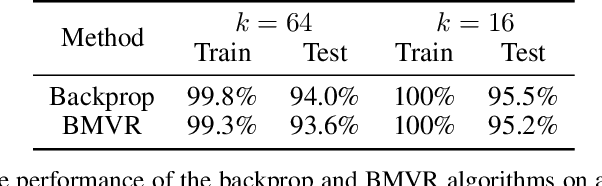
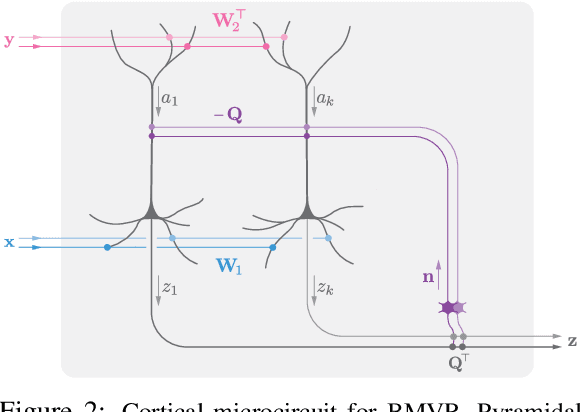
Abstract:The backpropagation algorithm is an invaluable tool for training artificial neural networks; however, because of a weight sharing requirement, it does not provide a plausible model of brain function. Here, in the context of a two-layer network, we derive an algorithm for training a neural network which avoids this problem by not requiring explicit error computation and backpropagation. Furthermore, our algorithm maps onto a neural network that bears a remarkable resemblance to the connectivity structure and learning rules of the cortex. We find that our algorithm empirically performs comparably to backprop on a number of datasets.
A simple normative network approximates local non-Hebbian learning in the cortex
Oct 23, 2020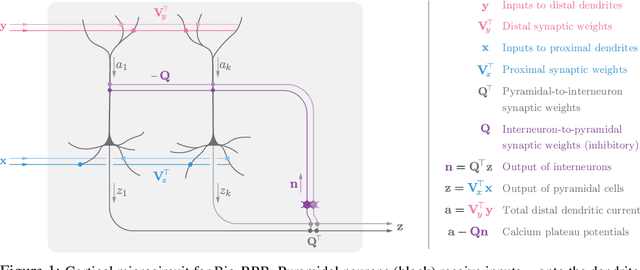
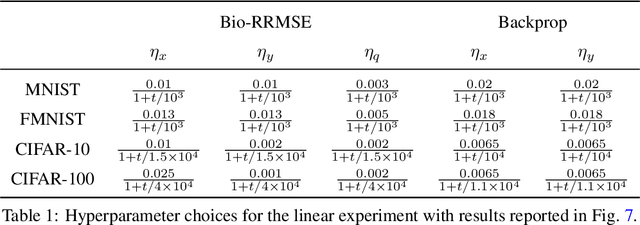
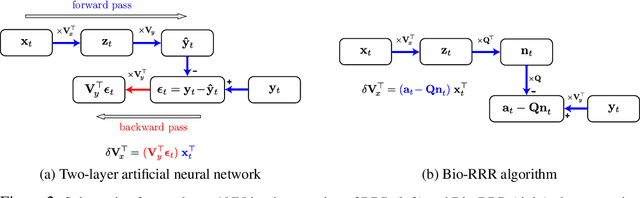
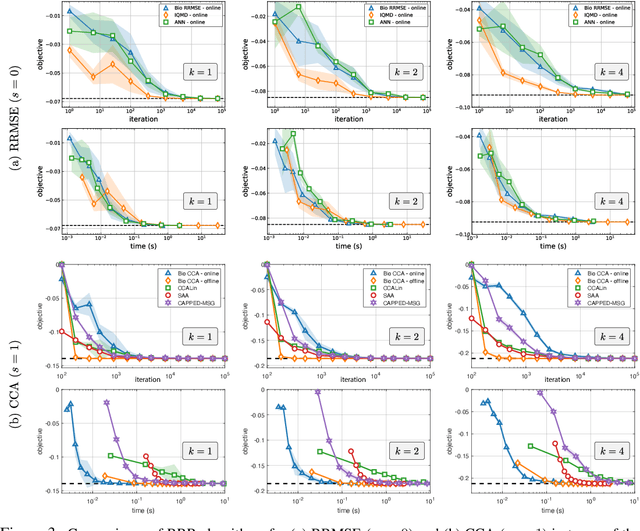
Abstract:To guide behavior, the brain extracts relevant features from high-dimensional data streamed by sensory organs. Neuroscience experiments demonstrate that the processing of sensory inputs by cortical neurons is modulated by instructive signals which provide context and task-relevant information. Here, adopting a normative approach, we model these instructive signals as supervisory inputs guiding the projection of the feedforward data. Mathematically, we start with a family of Reduced-Rank Regression (RRR) objective functions which include Reduced Rank (minimum) Mean Square Error (RRMSE) and Canonical Correlation Analysis (CCA), and derive novel offline and online optimization algorithms, which we call Bio-RRR. The online algorithms can be implemented by neural networks whose synaptic learning rules resemble calcium plateau potential dependent plasticity observed in the cortex. We detail how, in our model, the calcium plateau potential can be interpreted as a backpropagating error signal. We demonstrate that, despite relying exclusively on biologically plausible local learning rules, our algorithms perform competitively with existing implementations of RRMSE and CCA.
A biologically plausible neural network for Slow Feature Analysis
Oct 23, 2020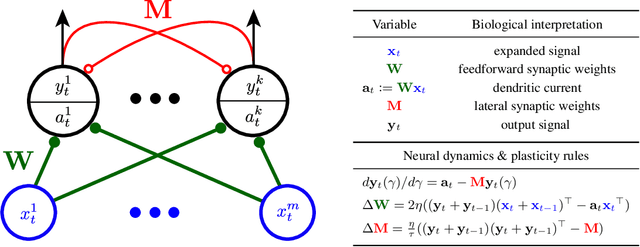
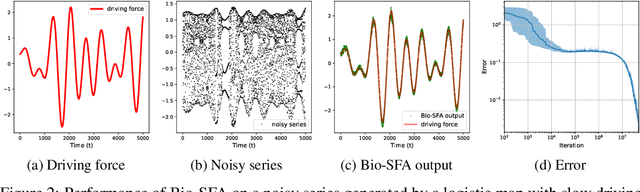
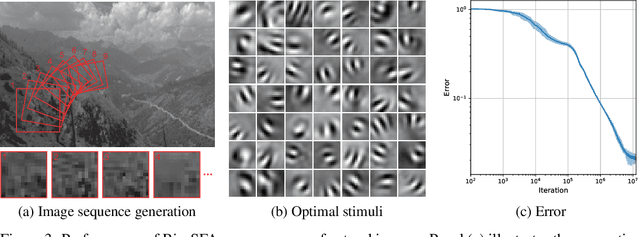
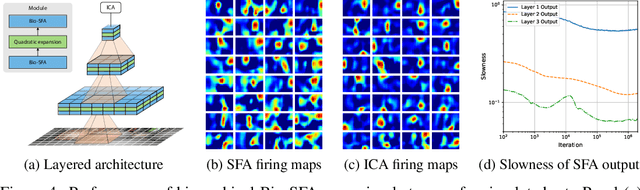
Abstract:Learning latent features from time series data is an important problem in both machine learning and brain function. One approach, called Slow Feature Analysis (SFA), leverages the slowness of many salient features relative to the rapidly varying input signals. Furthermore, when trained on naturalistic stimuli, SFA reproduces interesting properties of cells in the primary visual cortex and hippocampus, suggesting that the brain uses temporal slowness as a computational principle for learning latent features. However, despite the potential relevance of SFA for modeling brain function, there is currently no SFA algorithm with a biologically plausible neural network implementation, by which we mean an algorithm operates in the online setting and can be mapped onto a neural network with local synaptic updates. In this work, starting from an SFA objective, we derive an SFA algorithm, called Bio-SFA, with a biologically plausible neural network implementation. We validate Bio-SFA on naturalistic stimuli.
Bio-NICA: A biologically inspired single-layer network for Nonnegative Independent Component Analysis
Oct 23, 2020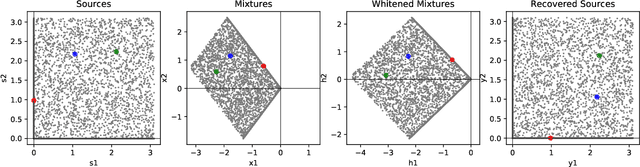
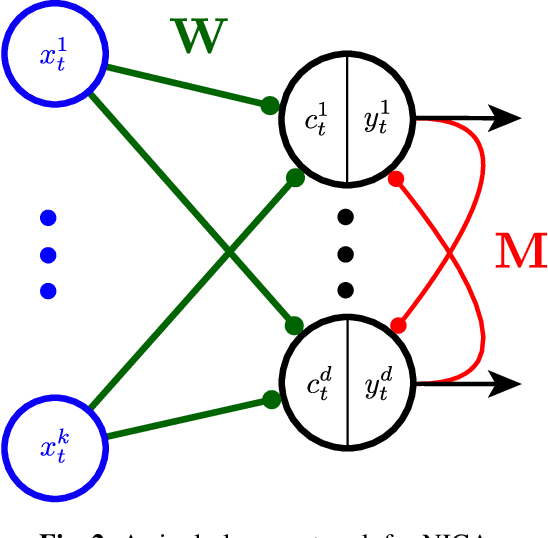
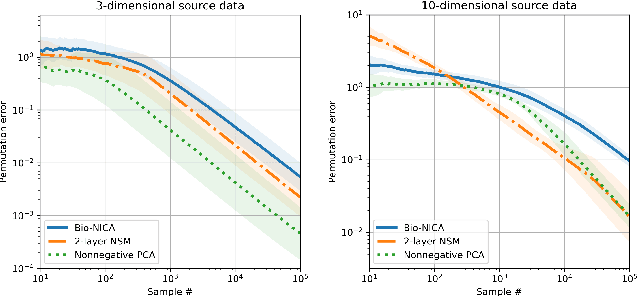

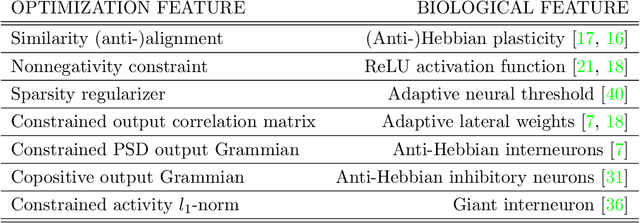
Abstract:Blind source separation, the problem of separating mixtures of unknown signals into their distinct sources, is an important problem for both biological and engineered signal processing systems. Nonnegative Independent Component Analysis (NICA) is a special case of blind source separation that assumes the mixture is a linear combination of independent, nonnegative sources. In this work, we derive a single-layer neural network implementation of NICA satisfying the following 3 constraints, which are relevant for biological systems and the design of neuromorphic hardware: (i) the network operates in the online setting, (ii) the synaptic learning rules are local, and (iii) the neural outputs are nonnegative.
Neuroscience-inspired online unsupervised learning algorithms
Sep 06, 2019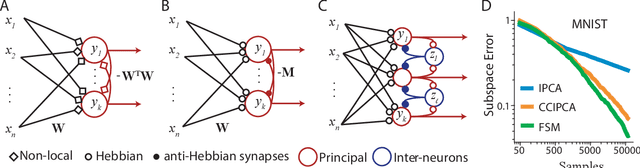
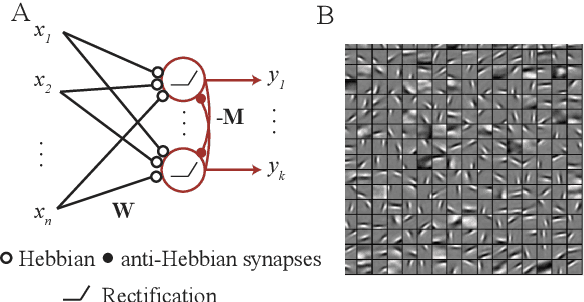
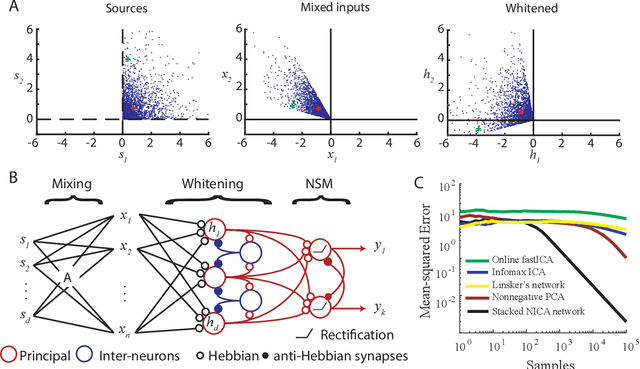
Abstract:Although the currently popular deep learning networks achieve unprecedented performance on some tasks, the human brain still has a monopoly on general intelligence. Motivated by this and biological implausibility of deep learning networks, we developed a family of biologically plausible artificial neural networks (NNs) for unsupervised learning. Our approach is based on optimizing principled objective functions containing a term that matches the pairwise similarity of outputs to the similarity of inputs, hence the name - similarity-based. Gradient-based online optimization of such similarity-based objective functions can be implemented by NNs with biologically plausible local learning rules. Similarity-based cost functions and associated NNs solve unsupervised learning tasks such as linear dimensionality reduction, sparse and/or nonnegative feature extraction, blind nonnegative source separation, clustering and manifold learning.
Efficient Principal Subspace Projection of Streaming Data Through Fast Similarity Matching
Aug 06, 2018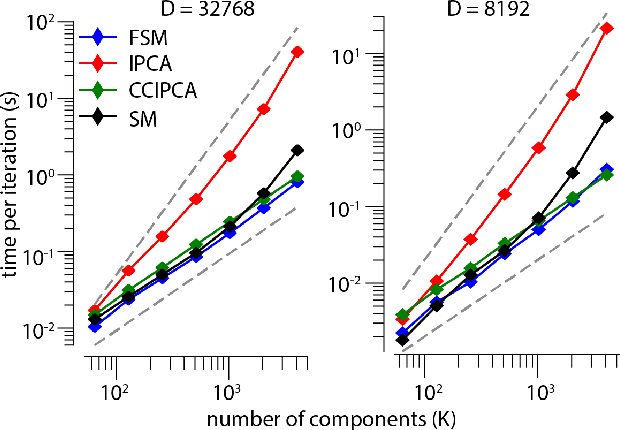
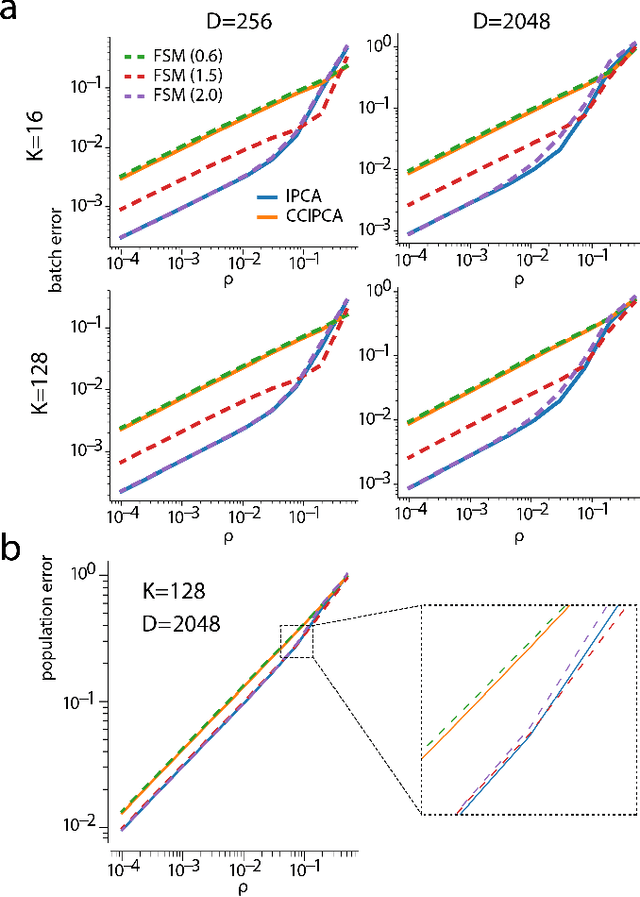
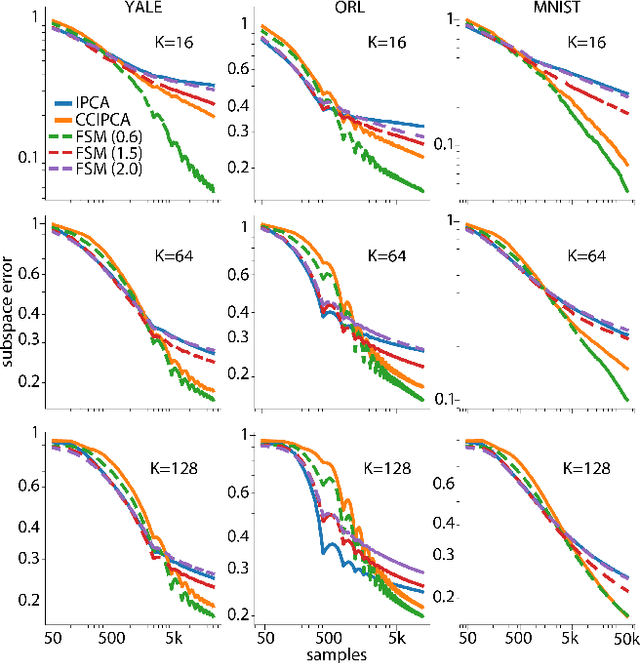
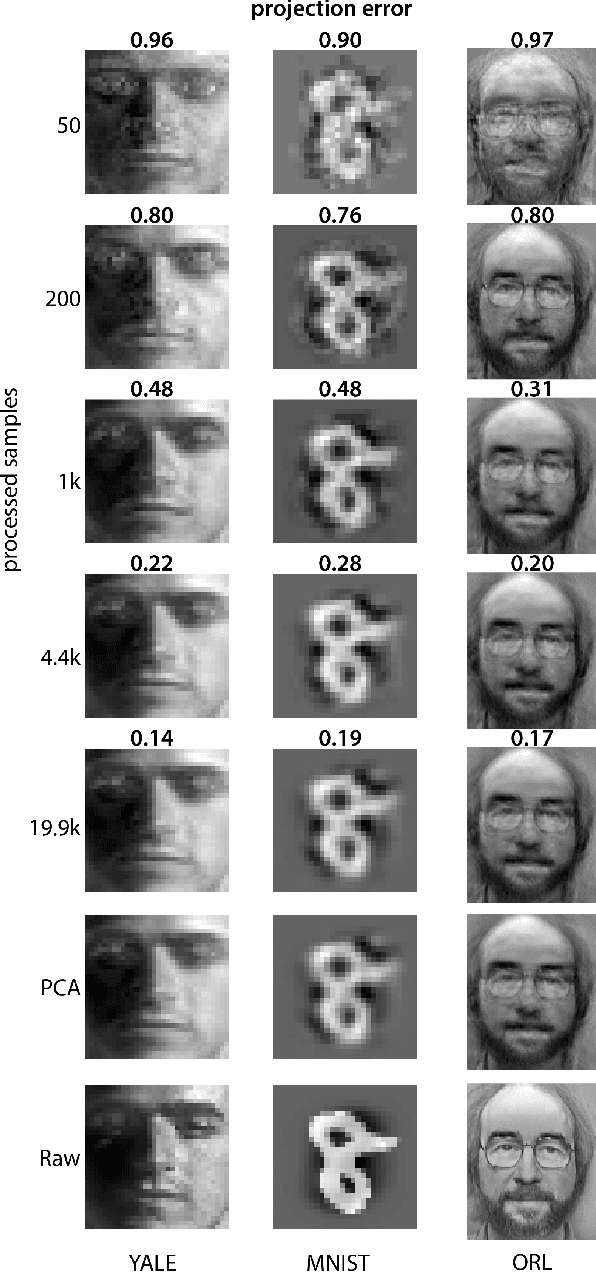
Abstract:Big data problems frequently require processing datasets in a streaming fashion, either because all data are available at once but collectively are larger than available memory or because the data intrinsically arrive one data point at a time and must be processed online. Here, we introduce a computationally efficient version of similarity matching, a framework for online dimensionality reduction that incrementally estimates the top K-dimensional principal subspace of streamed data while keeping in memory only the last sample and the current iterate. To assess the performance of our approach, we construct and make public a test suite containing both a synthetic data generator and the infrastructure to test online dimensionality reduction algorithms on real datasets, as well as performant implementations of our algorithm and competing algorithms with similar aims. Among the algorithms considered we find our approach to be competitive, performing among the best on both synthetic and real data.
Why do similarity matching objectives lead to Hebbian/anti-Hebbian networks?
Jul 11, 2017Abstract:Modeling self-organization of neural networks for unsupervised learning using Hebbian and anti-Hebbian plasticity has a long history in neuroscience. Yet, derivations of single-layer networks with such local learning rules from principled optimization objectives became possible only recently, with the introduction of similarity matching objectives. What explains the success of similarity matching objectives in deriving neural networks with local learning rules? Here, using dimensionality reduction as an example, we introduce several variable substitutions that illuminate the success of similarity matching. We show that the full network objective may be optimized separately for each synapse using local learning rules both in the offline and online settings. We formalize the long-standing intuition of the rivalry between Hebbian and anti-Hebbian rules by formulating a min-max optimization problem. We introduce a novel dimensionality reduction objective using fractional matrix exponents. To illustrate the generality of our approach, we apply it to a novel formulation of dimensionality reduction combined with whitening. We confirm numerically that the networks with learning rules derived from principled objectives perform better than those with heuristic learning rules.
Blind nonnegative source separation using biological neural networks
Jun 01, 2017Abstract:Blind source separation, i.e. extraction of independent sources from a mixture, is an important problem for both artificial and natural signal processing. Here, we address a special case of this problem when sources (but not the mixing matrix) are known to be nonnegative, for example, due to the physical nature of the sources. We search for the solution to this problem that can be implemented using biologically plausible neural networks. Specifically, we consider the online setting where the dataset is streamed to a neural network. The novelty of our approach is that we formulate blind nonnegative source separation as a similarity matching problem and derive neural networks from the similarity matching objective. Importantly, synaptic weights in our networks are updated according to biologically plausible local learning rules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge