David Thwaites
Institute of Medical Physics, School of Physics, University of Sydney, Sydney, New South Wales, Australia
Exploring Federated Deep Learning for Standardising Naming Conventions in Radiotherapy Data
Feb 14, 2024



Abstract:Standardising structure volume names in radiotherapy (RT) data is necessary to enable data mining and analyses, especially across multi-institutional centres. This process is time and resource intensive, which highlights the need for new automated and efficient approaches to handle the task. Several machine learning-based methods have been proposed and evaluated to standardise nomenclature. However, no studies have considered that RT patient records are distributed across multiple data centres. This paper introduces a method that emulates real-world environments to establish standardised nomenclature. This is achieved by integrating decentralised real-time data and federated learning (FL). A multimodal deep artificial neural network was proposed to standardise RT data in federated settings. Three types of possible attributes were extracted from the structures to train the deep learning models: tabular, visual, and volumetric. Simulated experiments were carried out to train the models across several scenarios including multiple data centres, input modalities, and aggregation strategies. The models were compared against models developed with single modalities in federated settings, in addition to models trained in centralised settings. Categorical classification accuracy was calculated on hold-out samples to inform the models performance. Our results highlight the need for fusing multiple modalities when training such models, with better performance reported with tabular-volumetric models. In addition, we report comparable accuracy compared to models built in centralised settings. This demonstrates the suitability of FL for handling the standardization task. Additional ablation analyses showed that the total number of samples in the data centres and the number of data centres highly affects the training process and should be carefully considered when building standardisation models.
Distributed learning optimisation of Cox models can leak patient data: Risks and solutions
Apr 12, 2022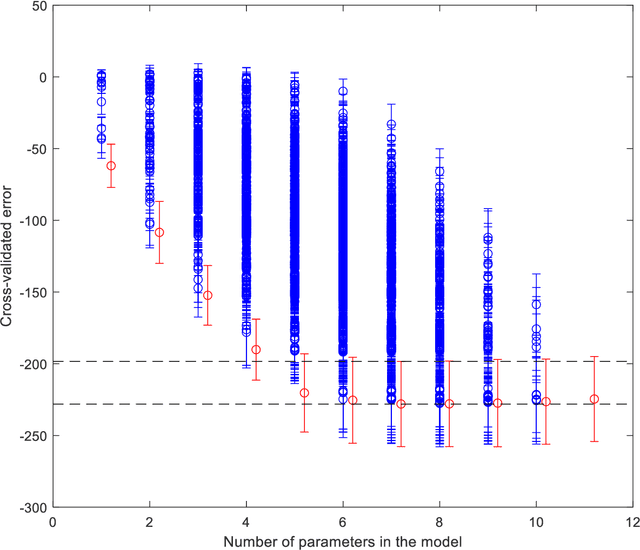
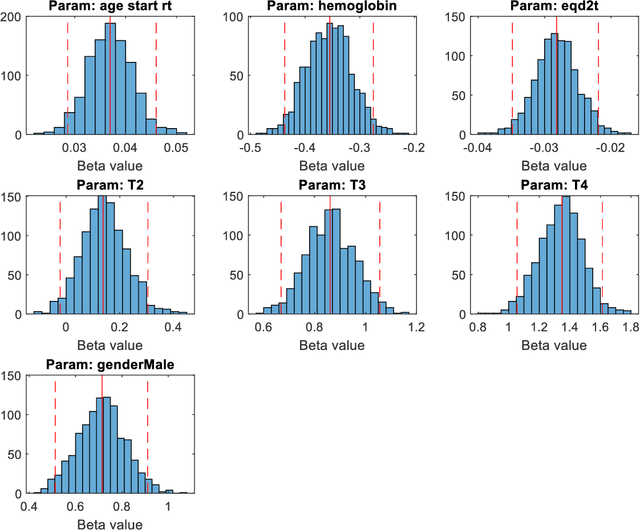
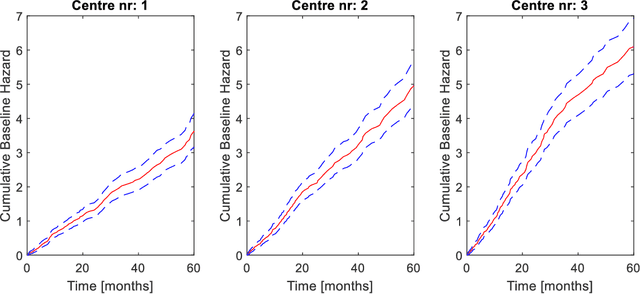
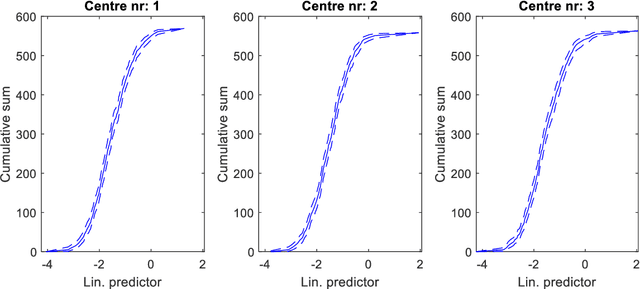
Abstract:Medical data are often highly sensitive, and frequently there are missing data. Due to the data's sensitive nature, there is an interest in creating modelling methods where the data are kept in each local centre to preserve their privacy, but yet the model can be trained on and learn from data across multiple centres. Such an approach might be distributed machine learning (federated learning, collaborative learning) in which a model is iteratively calculated based on aggregated local model information from each centre. However, even though no specific data are leaving the centre, there is a potential risk that the exchanged information is sufficient to reconstruct all or part of the patient data, which would hamper the safety-protecting rationale idea of distributed learning. This paper demonstrates that the optimisation of a Cox survival model can lead to patient data leakage. Following this, we suggest a way to optimise and validate a Cox model that avoids these problems in a secure way. The feasibility of the suggested method is demonstrated in a provided Matlab code that also includes methods for handling missing data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge