Collin Beaudoin
Quality Model for Machine Learning Components
Feb 04, 2026Abstract:Despite increased adoption and advances in machine learning (ML), there are studies showing that many ML prototypes do not reach the production stage and that testing is still largely limited to testing model properties, such as model performance, without considering requirements derived from the system it will be a part of, such as throughput, resource consumption, or robustness. This limited view of testing leads to failures in model integration, deployment, and operations. In traditional software development, quality models such as ISO 25010 provide a widely used structured framework to assess software quality, define quality requirements, and provide a common language for communication with stakeholders. A newer standard, ISO 25059, defines a more specific quality model for AI systems. However, a problem with this standard is that it combines system attributes with ML component attributes, which is not helpful for a model developer, as many system attributes cannot be assessed at the component level. In this paper, we present a quality model for ML components that serves as a guide for requirements elicitation and negotiation and provides a common vocabulary for ML component developers and system stakeholders to agree on and define system-derived requirements and focus their testing efforts accordingly. The quality model was validated through a survey in which the participants agreed with its relevance and value. The quality model has been successfully integrated into an open-source tool for ML component testing and evaluation demonstrating its practical application.
Q-Fusion: Diffusing Quantum Circuits
Apr 29, 2025Abstract:Quantum computing holds great potential for solving socially relevant and computationally complex problems. Furthermore, quantum machine learning (QML) promises to rapidly improve our current machine learning capabilities. However, current noisy intermediate-scale quantum (NISQ) devices are constrained by limitations in the number of qubits and gate counts, which hinder their full capabilities. Furthermore, the design of quantum algorithms remains a laborious task, requiring significant domain expertise and time. Quantum Architecture Search (QAS) aims to streamline this process by automatically generating novel quantum circuits, reducing the need for manual intervention. In this paper, we propose a diffusion-based algorithm leveraging the LayerDAG framework to generate new quantum circuits. This method contrasts with other approaches that utilize large language models (LLMs), reinforcement learning (RL), variational autoencoders (VAE), and similar techniques. Our results demonstrate that the proposed model consistently generates 100% valid quantum circuit outputs.
Evaluating Effects of Augmented SELFIES for Molecular Understanding Using QK-LSTM
Apr 29, 2025Abstract:Identifying molecular properties, including side effects, is a critical yet time-consuming step in drug development. Failing to detect these side effects before regulatory submission can result in significant financial losses and production delays, and overlooking them during the regulatory review can lead to catastrophic consequences. This challenge presents an opportunity for innovative machine learning approaches, particularly hybrid quantum-classical models like the Quantum Kernel-Based Long Short-Term Memory (QK-LSTM) network. The QK-LSTM integrates quantum kernel functions into the classical LSTM framework, enabling the capture of complex, non-linear patterns in sequential data. By mapping input data into a high-dimensional quantum feature space, the QK-LSTM model reduces the need for large parameter sets, allowing for model compression without sacrificing accuracy in sequence-based tasks. Recent advancements have been made in the classical domain using augmented variations of the Simplified Molecular Line-Entry System (SMILES). However, to the best of our knowledge, no research has explored the impact of augmented SMILES in the quantum domain, nor the role of augmented Self-Referencing Embedded Strings (SELFIES) in either classical or hybrid quantum-classical settings. This study presents the first analysis of these approaches, providing novel insights into their potential for enhancing molecular property prediction and side effect identification. Results reveal that augmenting SELFIES yields in statistically significant improvements from SMILES by a 5.97% improvement for the classical domain and a 5.91% improvement for the hybrid quantum-classical domain.
Predicting Side Effect of Drug Molecules using Recurrent Neural Networks
May 17, 2023Abstract:Identification and verification of molecular properties such as side effects is one of the most important and time-consuming steps in the process of molecule synthesis. For example, failure to identify side effects before submission to regulatory groups can cost millions of dollars and months of additional research to the companies. Failure to identify side effects during the regulatory review can also cost lives. The complexity and expense of this task have made it a candidate for a machine learning-based solution. Prior approaches rely on complex model designs and excessive parameter counts for side effect predictions. We believe reliance on complex models only shifts the difficulty away from chemists rather than alleviating the issue. Implementing large models is also expensive without prior access to high-performance computers. We propose a heuristic approach that allows for the utilization of simple neural networks, specifically the recurrent neural network, with a 98+% reduction in the number of required parameters compared to available large language models while still obtaining near identical results as top-performing models.
Energy-based Generative Models for Target-specific Drug Discovery
Dec 05, 2022Abstract:Drug targets are the main focus of drug discovery due to their key role in disease pathogenesis. Computational approaches are widely applied to drug development because of the increasing availability of biological molecular datasets. Popular generative approaches can create new drug molecules by learning the given molecule distributions. However, these approaches are mostly not for target-specific drug discovery. We developed an energy-based probabilistic model for computational target-specific drug discovery. Results show that our proposed TagMol can generate molecules with similar binding affinity scores as real molecules. GAT-based models showed faster and better learning relative to GCN baseline models.
Quantum Machine Learning for Material Synthesis and Hardware Security
Aug 16, 2022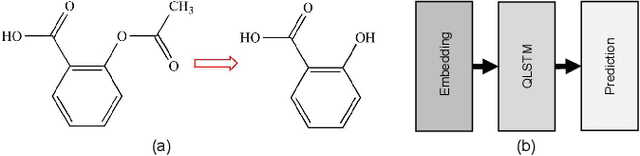
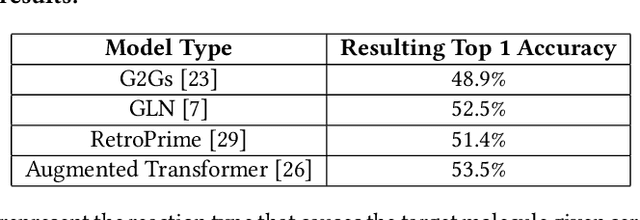
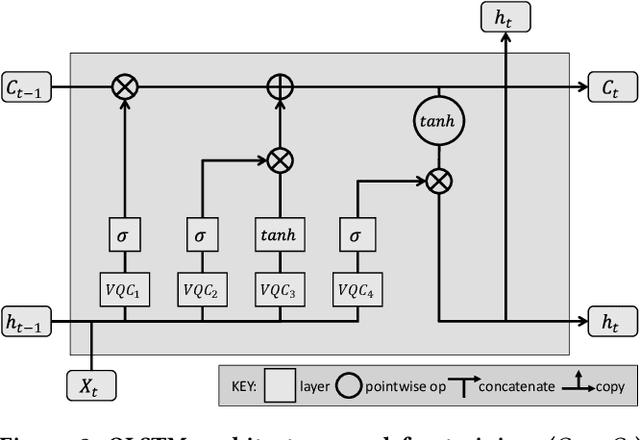

Abstract:Using quantum computing, this paper addresses two scientifically pressing and day-to-day relevant problems, namely, chemical retrosynthesis which is an important step in drug/material discovery and security of the semiconductor supply chain. We show that Quantum Long Short-Term Memory (QLSTM) is a viable tool for retrosynthesis. We achieve 65% training accuracy with QLSTM, whereas classical LSTM can achieve 100%. However, in testing, we achieve 80% accuracy with the QLSTM while classical LSTM peaks at only 70% accuracy! We also demonstrate an application of Quantum Neural Network (QNN) in the hardware security domain, specifically in Hardware Trojan (HT) detection using a set of power and area Trojan features. The QNN model achieves detection accuracy as high as 97.27%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge