Christian Etmann
on behalf of the AIX-COVNET collaboration
A Closer Look at Double Backpropagation
Jun 16, 2019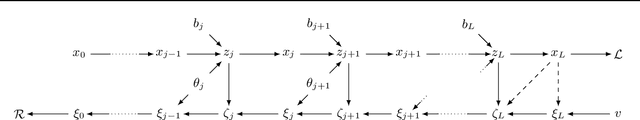
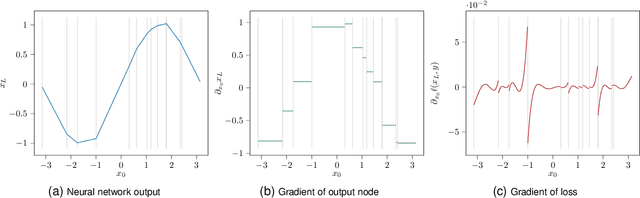
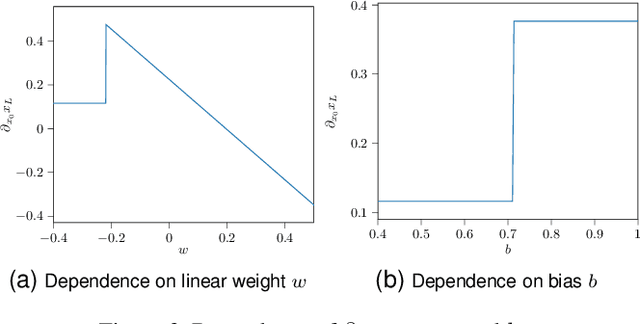
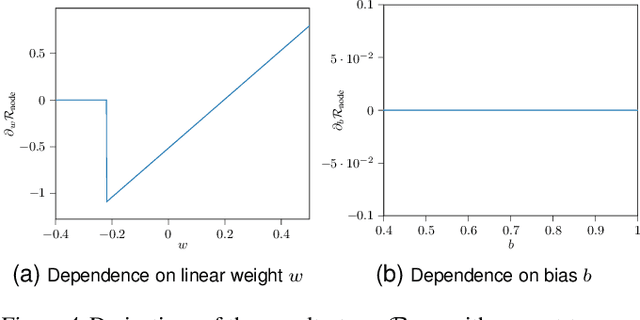
Abstract:In recent years, an increasing number of neural network models have included derivatives with respect to inputs in their loss functions, resulting in so-called double backpropagation for first-order optimization. However, so far no general description of the involved derivatives exists. Here, we cover a wide array of special cases in a very general Hilbert space framework, which allows us to provide optimized backpropagation rules for many real-world scenarios. This includes the reduction of calculations for Frobenius-norm-penalties on Jacobians by roughly a third for locally linear activation functions. Furthermore, we provide a description of the discontinuous loss surface of ReLU networks both in the inputs and the parameters and demonstrate why the discontinuities do not pose a big problem in reality.
On the Connection Between Adversarial Robustness and Saliency Map Interpretability
May 10, 2019
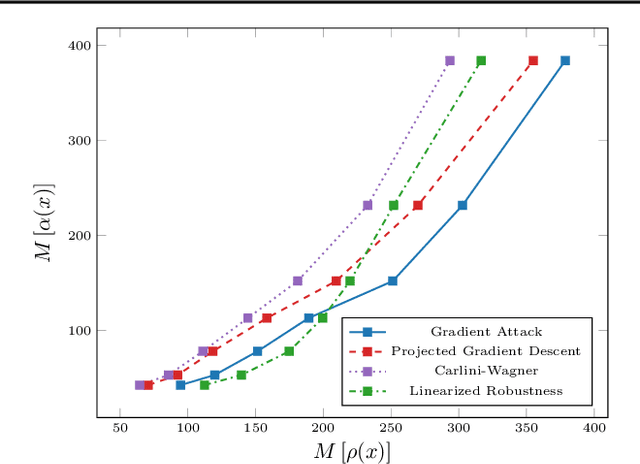
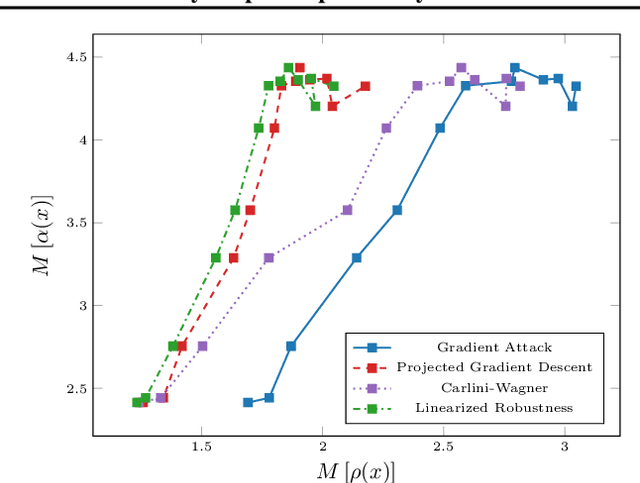
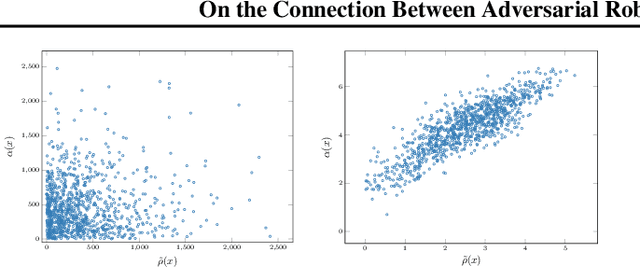
Abstract:Recent studies on the adversarial vulnerability of neural networks have shown that models trained to be more robust to adversarial attacks exhibit more interpretable saliency maps than their non-robust counterparts. We aim to quantify this behavior by considering the alignment between input image and saliency map. We hypothesize that as the distance to the decision boundary grows,so does the alignment. This connection is strictly true in the case of linear models. We confirm these theoretical findings with experiments based on models trained with a local Lipschitz regularization and identify where the non-linear nature of neural networks weakens the relation.
Deep Learning for Tumor Classification in Imaging Mass Spectrometry
Oct 20, 2017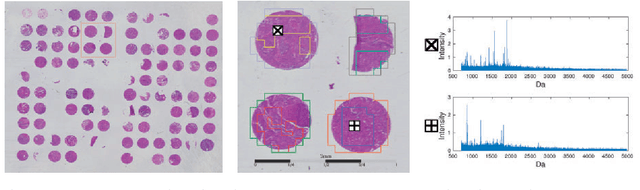
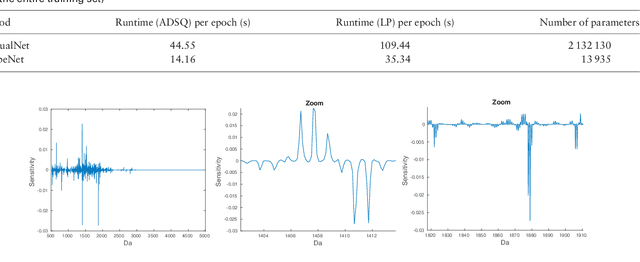
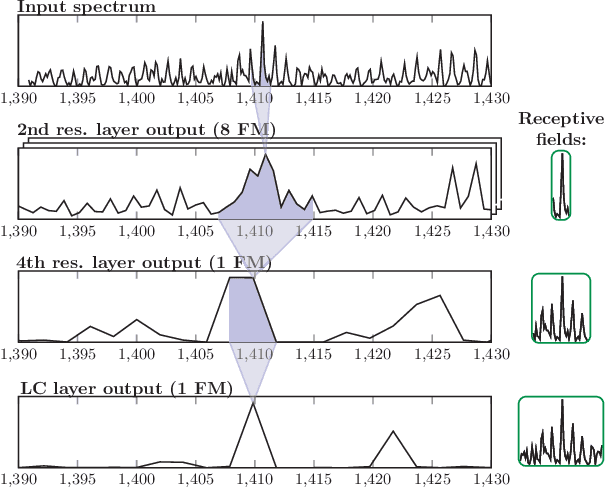
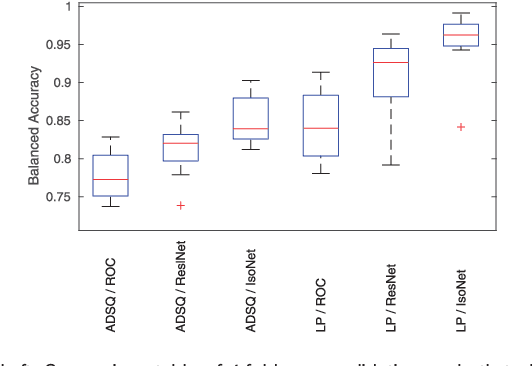
Abstract:Motivation: Tumor classification using Imaging Mass Spectrometry (IMS) data has a high potential for future applications in pathology. Due to the complexity and size of the data, automated feature extraction and classification steps are required to fully process the data. Deep learning offers an approach to learn feature extraction and classification combined in a single model. Commonly these steps are handled separately in IMS data analysis, hence deep learning offers an alternative strategy worthwhile to explore. Results: Methodologically, we propose an adapted architecture based on deep convolutional networks to handle the characteristics of mass spectrometry data, as well as a strategy to interpret the learned model in the spectral domain based on a sensitivity analysis. The proposed methods are evaluated on two challenging tumor classification tasks and compared to a baseline approach. Competitiveness of the proposed methods are shown on both tasks by studying the performance via cross-validation. Moreover, the learned models are analyzed by the proposed sensitivity analysis revealing biologically plausible effects as well as confounding factors of the considered task. Thus, this study may serve as a starting point for further development of deep learning approaches in IMS classification tasks.
* 10 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge