Chenyu Wen
Denoise Stepwise Signals by Diffusion Model Based Approach
Feb 09, 2026Abstract:Stepwise signals are ubiquitous in single-molecule detections, where abrupt changes in signal levels typically correspond to molecular conformational changes or state transitions. However, these features are inevitably obscured by noise, leading to uncertainty in estimating both signal levels and transition points. Traditional frequency-domain filtering is ineffective for denoising stepwise signals, as edge-related high-frequency components strongly overlap with noise. Although Hidden Markov Model-based approaches are widely used, they rely on stationarity assumptions and are not specifically designed for signal denoising. Here, we propose a diffusion model-based algorithm for stepwise signal denoising, named the Stepwise Signal Diffusion Model (SSDM). During training, SSDM learns the statistical structure of stepwise signals via a forward diffusion process that progressively adds noise. In the following reverse process, the model reconstructs clean signals from noisy observations, integrating a multi-scale convolutional network with an attention mechanism. Training data are generated by simulating stepwise signals through a Markov process with additive Gaussian noise. Across a broad range of signal-to-noise ratios, SSDM consistently outperforms traditional methods in both signal level reconstruction and transition point detection. Its effectiveness is further demonstrated on experimental data from single-molecule Forster Resonance Energy Transfer and nanopore DNA translocation measurements. Overall, SSDM provides a general and robust framework for recovering stepwise signals in various single-molecule detections and other physical systems exhibiting discrete state transitions.
Deep learning of nanopore sensing signals using a bi-path network
May 08, 2021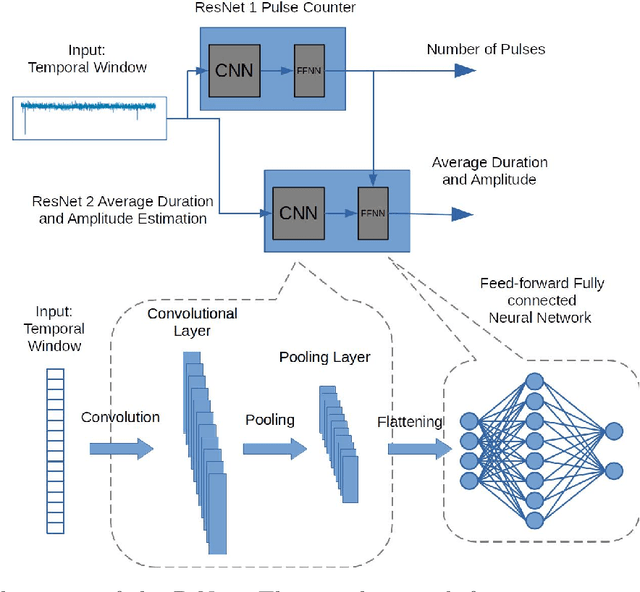
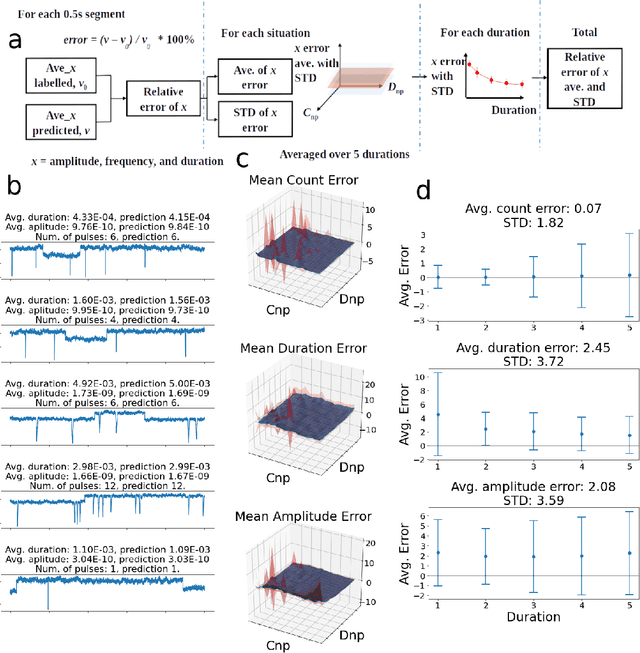
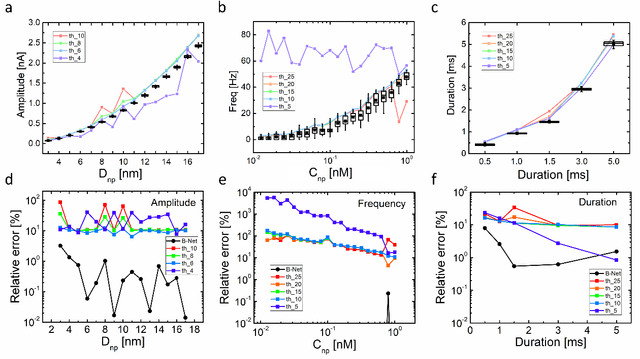
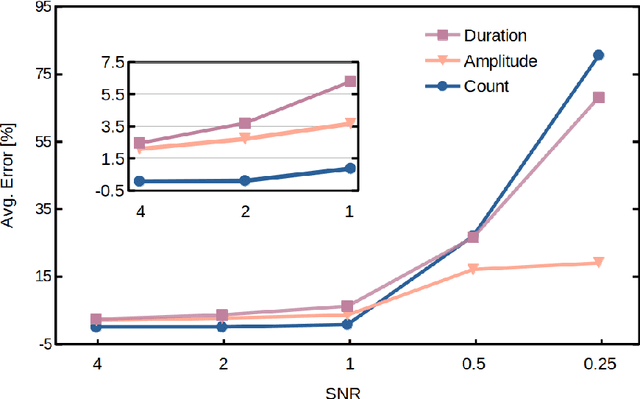
Abstract:Temporary changes in electrical resistance of a nanopore sensor caused by translocating target analytes are recorded as a sequence of pulses on current traces. Prevalent algorithms for feature extraction in pulse-like signals lack objectivity because empirical amplitude thresholds are user-defined to single out the pulses from the noisy background. Here, we use deep learning for feature extraction based on a bi-path network (B-Net). After training, the B-Net acquires the prototypical pulses and the ability of both pulse recognition and feature extraction without a priori assigned parameters. The B-Net performance is evaluated on generated datasets and further applied to experimental data of DNA and protein translocation. The B-Net results show remarkably small relative errors and stable trends. The B-Net is further shown capable of processing data with a signal-to-noise ratio equal to one, an impossibility for threshold-based algorithms. The developed B-Net is generic for pulse-like signals beyond pulsed nanopore currents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge