Cátia Pesquita
A Systematic Evaluation of Knowledge Graph Embeddings for Gene-Disease Association Prediction
Apr 11, 2025Abstract:Discovery gene-disease links is important in biology and medicine areas, enabling disease identification and drug repurposing. Machine learning approaches accelerate this process by leveraging biological knowledge represented in ontologies and the structure of knowledge graphs. Still, many existing works overlook ontologies explicitly representing diseases, missing causal and semantic relationships between them. The gene-disease association problem naturally frames itself as a link prediction task, where embedding algorithms directly predict associations by exploring the structure and properties of the knowledge graph. Some works frame it as a node-pair classification task, combining embedding algorithms with traditional machine learning algorithms. This strategy aligns with the logic of a machine learning pipeline. However, the use of negative examples and the lack of validated gene-disease associations to train embedding models may constrain its effectiveness. This work introduces a novel framework for comparing the performance of link prediction versus node-pair classification tasks, analyses the performance of state of the art gene-disease association approaches, and compares the different order-based formalizations of gene-disease association prediction. It also evaluates the impact of the semantic richness through a disease-specific ontology and additional links between ontologies. The framework involves five steps: data splitting, knowledge graph integration, embedding, modeling and prediction, and method evaluation. Results show that enriching the semantic representation of diseases slightly improves performance, while additional links generate a greater impact. Link prediction methods better explore the semantic richness encoded in knowledge graphs. Although node-pair classification methods identify all true positives, link prediction methods outperform overall.
Ontology alignment repair through modularization and confidence-based heuristics
Jul 19, 2013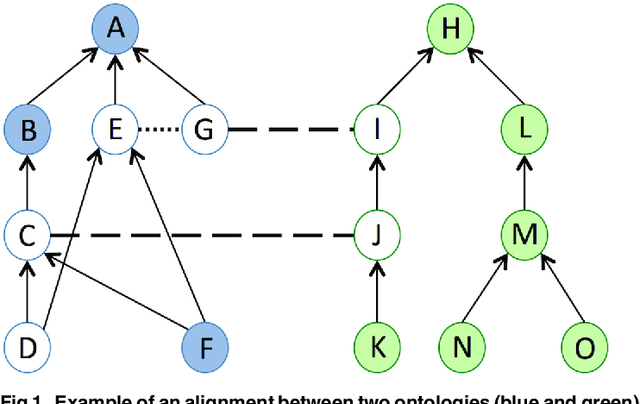
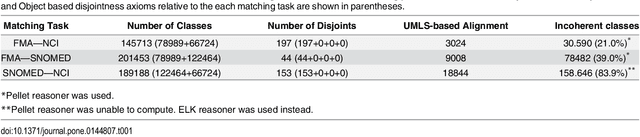
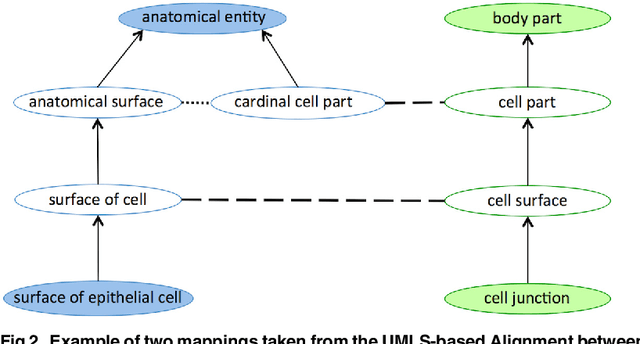
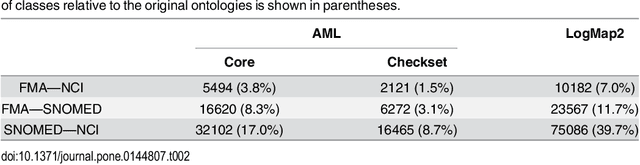
Abstract:Ontology Matching aims to find a set of semantic correspondences, called an alignment, between related ontologies. In recent years, there has been a growing interest in efficient and effective matching methods for large ontologies. However, most of the alignments produced for large ontologies are logically incoherent. It was only recently that the use of repair techniques to improve the quality of ontology alignments has been explored. In this paper we present a novel technique for detecting incoherent concepts based on ontology modularization, and a new repair algorithm that minimizes the incoherence of the resulting alignment and the number of matches removed from the input alignment. An implementation was done as part of a lightweight version of AgreementMaker system, a successful ontology matching platform, and evaluated using a set of four benchmark biomedical ontology matching tasks. Our results show that our implementation is efficient and produces better alignments with respect to their coherence and f-measure than the state of the art repairing tools. They also show that our implementation is a better alternative for producing coherent silver standard alignments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge