Ata Kiapour
A simple and effective approach for body part recognition on CT scans based on projection estimation
Apr 30, 2025Abstract:It is well known that machine learning models require a high amount of annotated data to obtain optimal performance. Labelling Computed Tomography (CT) data can be a particularly challenging task due to its volumetric nature and often missing and$/$or incomplete associated meta-data. Even inspecting one CT scan requires additional computer software, or in the case of programming languages $-$ additional programming libraries. This study proposes a simple, yet effective approach based on 2D X-ray-like estimation of 3D CT scans for body region identification. Although body region is commonly associated with the CT scan, it often describes only the focused major body region neglecting other anatomical regions present in the observed CT. In the proposed approach, estimated 2D images were utilized to identify 14 distinct body regions, providing valuable information for constructing a high-quality medical dataset. To evaluate the effectiveness of the proposed method, it was compared against 2.5D, 3D and foundation model (MI2) based approaches. Our approach outperformed the others, where it came on top with statistical significance and F1-Score for the best-performing model EffNet-B0 of 0.980 $\pm$ 0.016 in comparison to the 0.840 $\pm$ 0.114 (2.5D DenseNet-161), 0.854 $\pm$ 0.096 (3D VoxCNN), and 0.852 $\pm$ 0.104 (MI2 foundation model). The utilized dataset comprised three different clinical centers and counted 15,622 CT scans (44,135 labels).
Do not Mask Randomly: Effective Domain-adaptive Pre-training by Masking In-domain Keywords
Jul 14, 2023



Abstract:We propose a novel task-agnostic in-domain pre-training method that sits between generic pre-training and fine-tuning. Our approach selectively masks in-domain keywords, i.e., words that provide a compact representation of the target domain. We identify such keywords using KeyBERT (Grootendorst, 2020). We evaluate our approach using six different settings: three datasets combined with two distinct pre-trained language models (PLMs). Our results reveal that the fine-tuned PLMs adapted using our in-domain pre-training strategy outperform PLMs that used in-domain pre-training with random masking as well as those that followed the common pre-train-then-fine-tune paradigm. Further, the overhead of identifying in-domain keywords is reasonable, e.g., 7-15% of the pre-training time (for two epochs) for BERT Large (Devlin et al., 2019).
A Compact Pretraining Approach for Neural Language Models
Aug 29, 2022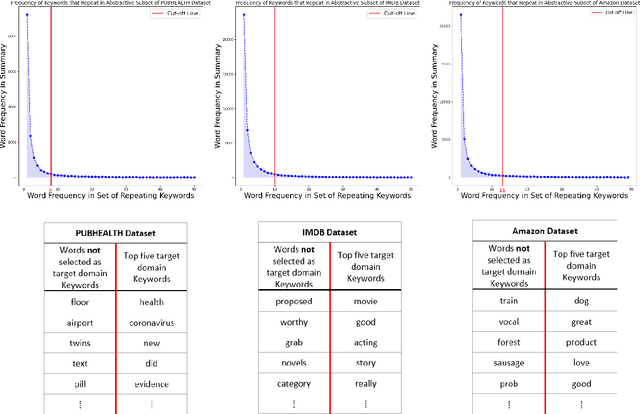
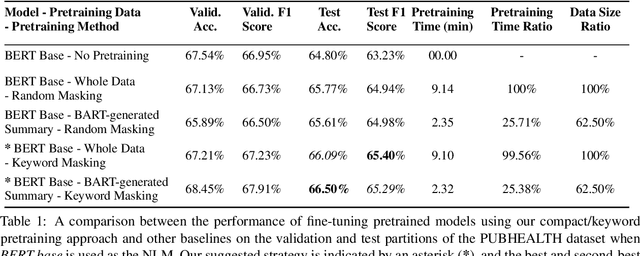
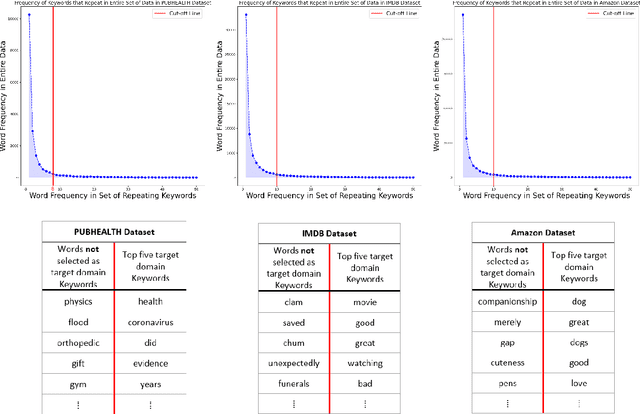
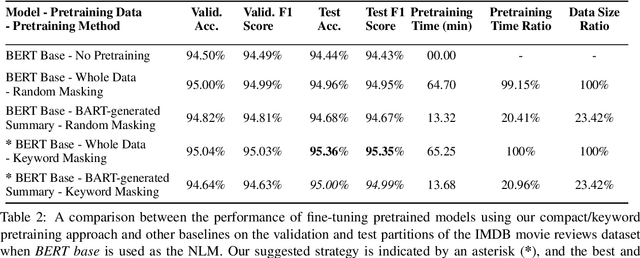
Abstract:Domain adaptation for large neural language models (NLMs) is coupled with massive amounts of unstructured data in the pretraining phase. In this study, however, we show that pretrained NLMs learn in-domain information more effectively and faster from a compact subset of the data that focuses on the key information in the domain. We construct these compact subsets from the unstructured data using a combination of abstractive summaries and extractive keywords. In particular, we rely on BART to generate abstractive summaries, and KeyBERT to extract keywords from these summaries (or the original unstructured text directly). We evaluate our approach using six different settings: three datasets combined with two distinct NLMs. Our results reveal that the task-specific classifiers trained on top of NLMs pretrained using our method outperform methods based on traditional pretraining, i.e., random masking on the entire data, as well as methods without pretraining. Further, we show that our strategy reduces pretraining time by up to five times compared to vanilla pretraining. The code for all of our experiments is publicly available at https://github.com/shahriargolchin/compact-pretraining.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge