Asif Rahman
MAD-OOD: A Deep Learning Cluster-Driven Framework for an Out-of-Distribution Malware Detection and Classification
Dec 19, 2025Abstract:Out of distribution (OOD) detection remains a critical challenge in malware classification due to the substantial intra family variability introduced by polymorphic and metamorphic malware variants. Most existing deep learning based malware detectors rely on closed world assumptions and fail to adequately model this intra class variation, resulting in degraded performance when confronted with previously unseen malware families. This paper presents MADOOD, a novel two stage, cluster driven deep learning framework for robust OOD malware detection and classification. In the first stage, malware family embeddings are modeled using class conditional spherical decision boundaries derived from Gaussian Discriminant Analysis (GDA), enabling statistically grounded separation of indistribution and OOD samples without requiring OOD data during training. Z score based distance analysis across multiple class centroids is employed to reliably identify anomalous samples in the latent space. In the second stage, a deep neural network integrates cluster based predictions, refined embeddings, and supervised classifier outputs to enhance final classification accuracy. Extensive evaluations on benchmark malware datasets comprising 25 known families and multiple novel OOD variants demonstrate that MADOOD significantly outperforms state of the art OOD detection methods, achieving an AUC of up to 0.911 on unseen malware families. The proposed framework provides a scalable, interpretable, and statistically principled solution for real world malware detection and anomaly identification in evolving cybersecurity environments.
MARCO: A Multi-Agent System for Optimizing HPC Code Generation Using Large Language Models
May 06, 2025Abstract:Large language models (LLMs) have transformed software development through code generation capabilities, yet their effectiveness for high-performance computing (HPC) remains limited. HPC code requires specialized optimizations for parallelism, memory efficiency, and architecture-specific considerations that general-purpose LLMs often overlook. We present MARCO (Multi-Agent Reactive Code Optimizer), a novel framework that enhances LLM-generated code for HPC through a specialized multi-agent architecture. MARCO employs separate agents for code generation and performance evaluation, connected by a feedback loop that progressively refines optimizations. A key innovation is MARCO's web-search component that retrieves real-time optimization techniques from recent conference proceedings and research publications, bridging the knowledge gap in pre-trained LLMs. Our extensive evaluation on the LeetCode 75 problem set demonstrates that MARCO achieves a 14.6% average runtime reduction compared to Claude 3.5 Sonnet alone, while the integration of the web-search component yields a 30.9% performance improvement over the base MARCO system. These results highlight the potential of multi-agent systems to address the specialized requirements of high-performance code generation, offering a cost-effective alternative to domain-specific model fine-tuning.
Interpretable Additive Recurrent Neural Networks For Multivariate Clinical Time Series
Sep 15, 2021Abstract:Time series models with recurrent neural networks (RNNs) can have high accuracy but are unfortunately difficult to interpret as a result of feature-interactions, temporal-interactions, and non-linear transformations. Interpretability is important in domains like healthcare where constructing models that provide insight into the relationships they have learned are required to validate and trust model predictions. We want accurate time series models where users can understand the contribution of individual input features. We present the Interpretable-RNN (I-RNN) that balances model complexity and accuracy by forcing the relationship between variables in the model to be additive. Interactions are restricted between hidden states of the RNN and additively combined at the final step. I-RNN specifically captures the unique characteristics of clinical time series, which are unevenly sampled in time, asynchronously acquired, and have missing data. Importantly, the hidden state activations represent feature coefficients that correlate with the prediction target and can be visualized as risk curves that capture the global relationship between individual input features and the outcome. We evaluate the I-RNN model on the Physionet 2012 Challenge dataset to predict in-hospital mortality, and on a real-world clinical decision support task: predicting hemodynamic interventions in the intensive care unit. I-RNN provides explanations in the form of global and local feature importances comparable to highly intelligible models like decision trees trained on hand-engineered features while significantly outperforming them. I-RNN remains intelligible while providing accuracy comparable to state-of-the-art decay-based and interpolation-based recurrent time series models. The experimental results on real-world clinical datasets refute the myth that there is a tradeoff between accuracy and interpretability.
Densely Connected Convolutional Networks and Signal Quality Analysis to Detect Atrial Fibrillation Using Short Single-Lead ECG Recordings
Oct 10, 2017
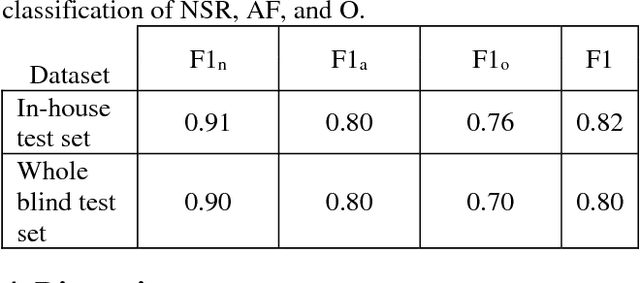
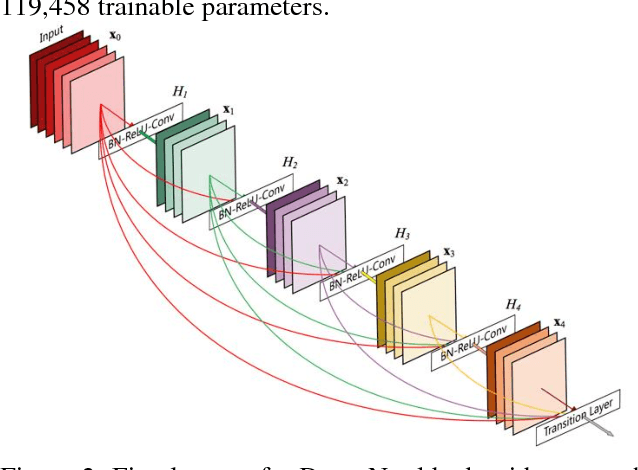
Abstract:The development of new technology such as wearables that record high-quality single channel ECG, provides an opportunity for ECG screening in a larger population, especially for atrial fibrillation screening. The main goal of this study is to develop an automatic classification algorithm for normal sinus rhythm (NSR), atrial fibrillation (AF), other rhythms (O), and noise from a single channel short ECG segment (9-60 seconds). For this purpose, signal quality index (SQI) along with dense convolutional neural networks was used. Two convolutional neural network (CNN) models (main model that accepts 15 seconds ECG and secondary model that processes 9 seconds shorter ECG) were trained using the training data set. If the recording is determined to be of low quality by SQI, it is immediately classified as noisy. Otherwise, it is transformed to a time-frequency representation and classified with the CNN as NSR, AF, O, or noise. At the final step, a feature-based post-processing algorithm classifies the rhythm as either NSR or O in case the CNN model's discrimination between the two is indeterminate. The best result achieved at the official phase of the PhysioNet/CinC challenge on the blind test set was 0.80 (F1 for NSR, AF, and O were 0.90, 0.80, and 0.70, respectively).
An Ensemble Boosting Model for Predicting Transfer to the Pediatric Intensive Care Unit
Jul 16, 2017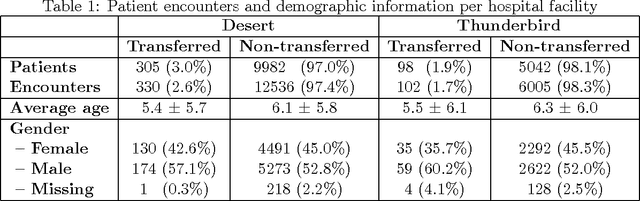
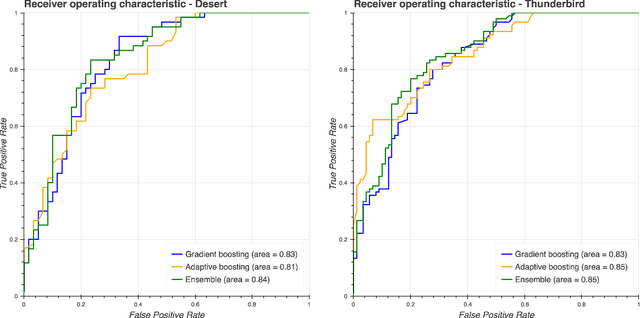
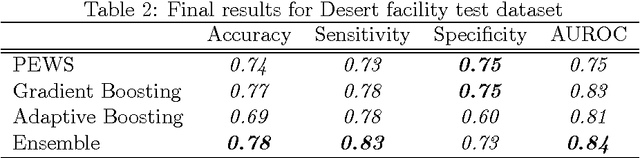
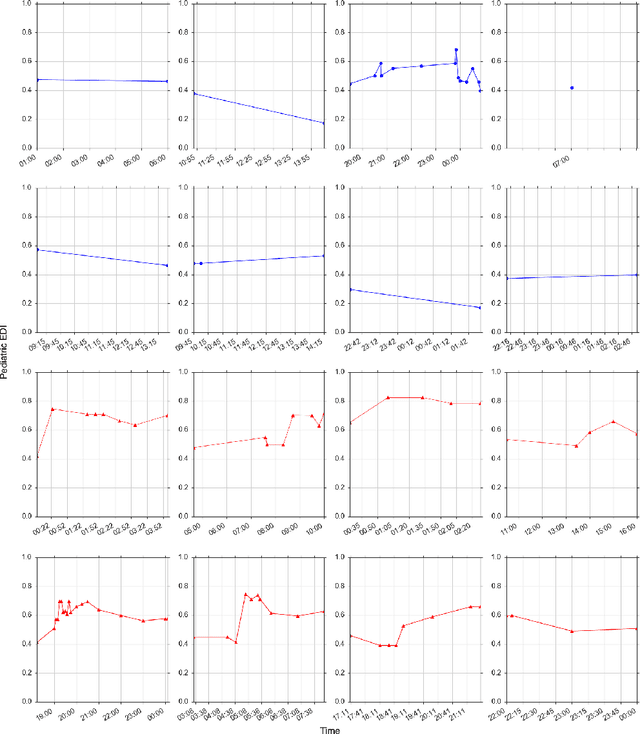
Abstract:Our work focuses on the problem of predicting the transfer of pediatric patients from the general ward of a hospital to the pediatric intensive care unit. Using data collected over 5.5 years from the electronic health records of two medical facilities, we develop classifiers based on adaptive boosting and gradient tree boosting. We further combine these learned classifiers into an ensemble model and compare its performance to a modified pediatric early warning score (PEWS) baseline that relies on expert defined guidelines. To gauge model generalizability, we perform an inter-facility evaluation where we train our algorithm on data from one facility and perform evaluation on a hidden test dataset from a separate facility. We show that improvements are witnessed over the PEWS baseline in accuracy (0.77 vs. 0.69), sensitivity (0.80 vs. 0.68), specificity (0.74 vs. 0.70) and AUROC (0.85 vs. 0.73).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge