Aras T. Asaad
A Novel Patch-Based TDA Approach for Computed Tomography
Dec 13, 2025



Abstract:The development of machine learning (ML) models based on computed tomography (CT) imaging modality has been a major focus of recent research in the medical imaging domain. Incorporating robust feature engineering approach can highly improve the performance of these models. Topological data analysis (TDA), a recent development based on the mathematical field of algebraic topology, mainly focuses on the data from a topological perspective, extracting deeper insight and higher dimensional structures from the data. Persistent homology (PH), a fundamental tool in the area of TDA, can extract topological features such as connected components, cycles and voids from the data. A popular approach to construct PH from 3D CT images is to utilize the 3D cubical complex filtration, a method adapted for grid-structured data. However, this approach may not always yield the best performance and can suffer from computational complexity with higher resolution CT images. This study introduces a novel patch-based PH construction approach tailored for volumetric medical imaging data, in particular CT modality. A wide range of experiments has been conducted on several datasets of 3D CT images to comprehensively analyze the performance of the proposed method with various parameters and benchmark it against the 3D cubical complex algorithm. Our results highlight the dominance of the patch-based TDA approach in terms of both classification performance and time-efficiency. The proposed approach outperformed the cubical complex method, achieving average improvement of 10.38%, 6.94%, 2.06%, 11.58%, and 8.51% in accuracy, AUC, sensitivity, specificity, and F1 score, respectively, across all datasets. Finally, we provide a convenient python package, Patch-TDA, to facilitate the utilization of the proposed approach.
Comparing the Effects of Persistence Barcodes Aggregation and Feature Concatenation on Medical Imaging
May 29, 2025



Abstract:In medical image analysis, feature engineering plays an important role in the design and performance of machine learning models. Persistent homology (PH), from the field of topological data analysis (TDA), demonstrates robustness and stability to data perturbations and addresses the limitation from traditional feature extraction approaches where a small change in input results in a large change in feature representation. Using PH, we store persistent topological and geometrical features in the form of the persistence barcode whereby large bars represent global topological features and small bars encapsulate geometrical information of the data. When multiple barcodes are computed from 2D or 3D medical images, two approaches can be used to construct the final topological feature vector in each dimension: aggregating persistence barcodes followed by featurization or concatenating topological feature vectors derived from each barcode. In this study, we conduct a comprehensive analysis across diverse medical imaging datasets to compare the effects of the two aforementioned approaches on the performance of classification models. The results of this analysis indicate that feature concatenation preserves detailed topological information from individual barcodes, yields better classification performance and is therefore a preferred approach when conducting similar experiments.
A Morse Transform for Drug Discovery
Mar 06, 2025Abstract:We introduce a new ligand-based virtual screening (LBVS) framework that uses piecewise linear (PL) Morse theory to predict ligand binding potential. We model ligands as simplicial complexes via a pruned Delaunay triangulation, and catalogue the critical points across multiple directional height functions. This produces a rich feature vector, consisting of crucial topological features -- peaks, troughs, and saddles -- that characterise ligand surfaces relevant to binding interactions. Unlike contemporary LBVS methods that rely on computationally-intensive deep neural networks, we require only a lightweight classifier. The Morse theoretic approach achieves state-of-the-art performance on standard datasets while offering an interpretable feature vector and scalable method for ligand prioritization in early-stage drug discovery.
NER- RoBERTa: Fine-Tuning RoBERTa for Named Entity Recognition (NER) within low-resource languages
Dec 15, 2024



Abstract:Nowadays, Natural Language Processing (NLP) is an important tool for most people's daily life routines, ranging from understanding speech, translation, named entity recognition (NER), and text categorization, to generative text models such as ChatGPT. Due to the existence of big data and consequently large corpora for widely used languages like English, Spanish, Turkish, Persian, and many more, these applications have been developed accurately. However, the Kurdish language still requires more corpora and large datasets to be included in NLP applications. This is because Kurdish has a rich linguistic structure, varied dialects, and a limited dataset, which poses unique challenges for Kurdish NLP (KNLP) application development. While several studies have been conducted in KNLP for various applications, Kurdish NER (KNER) remains a challenge for many KNLP tasks, including text analysis and classification. In this work, we address this limitation by proposing a methodology for fine-tuning the pre-trained RoBERTa model for KNER. To this end, we first create a Kurdish corpus, followed by designing a modified model architecture and implementing the training procedures. To evaluate the trained model, a set of experiments is conducted to demonstrate the performance of the KNER model using different tokenization methods and trained models. The experimental results show that fine-tuned RoBERTa with the SentencePiece tokenization method substantially improves KNER performance, achieving a 12.8% improvement in F1-score compared to traditional models, and consequently establishes a new benchmark for KNLP.
Diagnosing COVID-19 Pneumonia from X-Ray and CT Images using Deep Learning and Transfer Learning Algorithms
Mar 31, 2020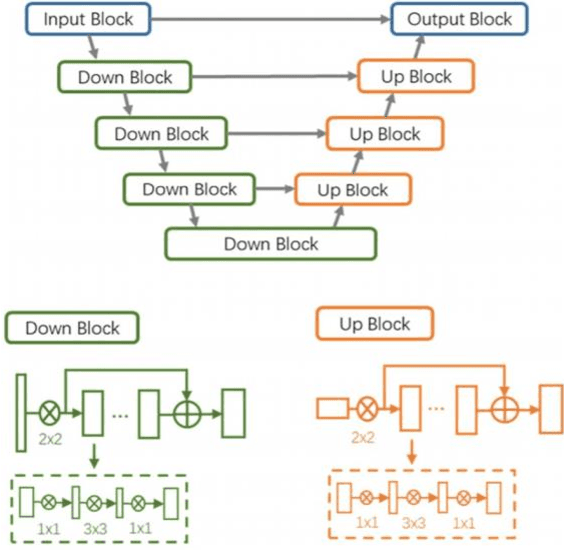
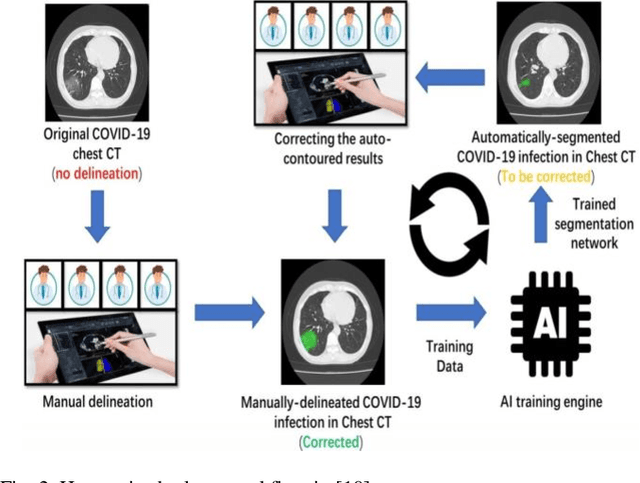
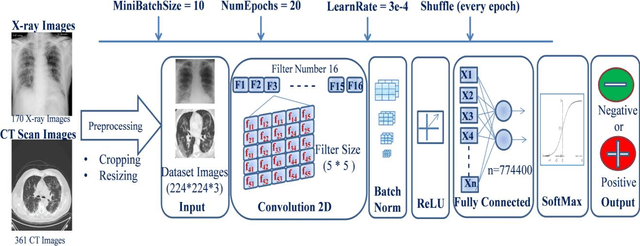
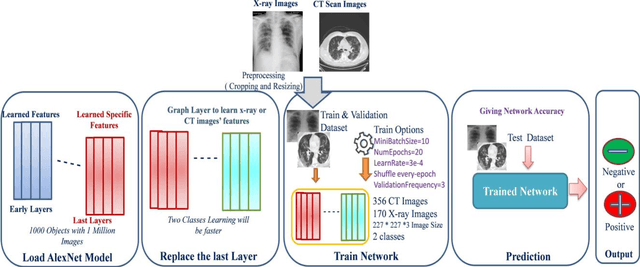
Abstract:COVID-19 (also known as 2019 Novel Coronavirus) first emerged in Wuhan, China and spread across the globe with unprecedented effect and has now become the greatest crisis of the modern era. The COVID-19 has proved much more pervasive demands for diagnosis that has driven researchers to develop more intelligent, highly responsive and efficient detection methods. In this work, we focus on proposing AI tools that can be used by radiologists or healthcare professionals to diagnose COVID-19 cases in a quick and accurate manner. However, the lack of a publicly available dataset of X-ray and CT images makes the design of such AI tools a challenging task. To this end, this study aims to build a comprehensive dataset of X-rays and CT scan images from multiple sources as well as provides a simple but an effective COVID-19 detection technique using deep learning and transfer learning algorithms. In this vein, a simple convolution neural network (CNN) and modified pre-trained AlexNet model are applied on the prepared X-rays and CT scan images dataset. The result of the experiments shows that the utilized models can provide accuracy up to 98 % via pre-trained network and 94.1 % accuracy by using the modified CNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge