Angel Alberich-Bayarri
Automated Machine Learning in Radiomics: A Comparative Evaluation of Performance, Efficiency and Accessibility
Jan 13, 2026Abstract:Automated machine learning (AutoML) frameworks can lower technical barriers for predictive and prognostic model development in radiomics by enabling researchers without programming expertise to build models. However, their effectiveness in addressing radiomics-specific challenges remains unclear. This study evaluates the performance, efficiency, and accessibility of general-purpose and radiomics-specific AutoML frameworks on diverse radiomics classification tasks, thereby highlighting development needs for radiomics. Ten public/private radiomics datasets with varied imaging modalities (CT/MRI), sizes, anatomies and endpoints were used. Six general-purpose and five radiomics-specific frameworks were tested with predefined parameters using standardized cross-validation. Evaluation metrics included AUC, runtime, together with qualitative aspects related to software status, accessibility, and interpretability. Simplatab, a radiomics-specific tool with a no-code interface, achieved the highest average test AUC (81.81%) with a moderate runtime (~1 hour). LightAutoML, a general-purpose framework, showed the fastest execution with competitive performance (78.74% mean AUC in six minutes). Most radiomics-specific frameworks were excluded from the performance analysis due to obsolescence, extensive programming requirements, or computational inefficiency. Conversely, general-purpose frameworks demonstrated higher accessibility and ease of implementation. Simplatab provides an effective balance of performance, efficiency, and accessibility for radiomics classification problems. However, significant gaps remain, including the lack of accessible survival analysis support and the limited integration of feature reproducibility and harmonization within current AutoML frameworks. Future research should focus on adapting AutoML solutions to better address these radiomics-specific challenges.
Data Harmonisation for Information Fusion in Digital Healthcare: A State-of-the-Art Systematic Review, Meta-Analysis and Future Research Directions
Jan 17, 2022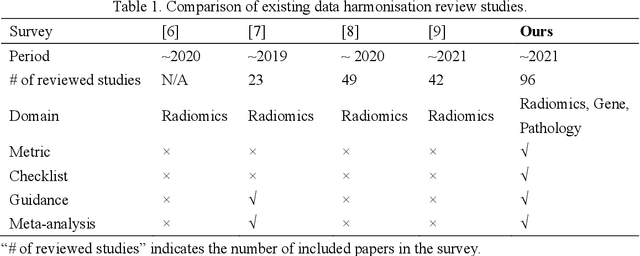

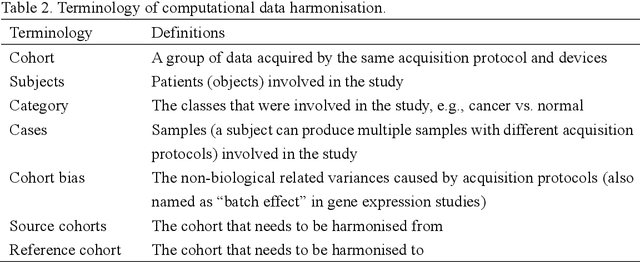
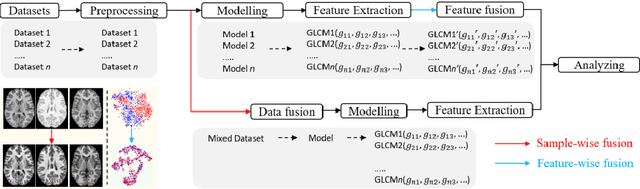
Abstract:Removing the bias and variance of multicentre data has always been a challenge in large scale digital healthcare studies, which requires the ability to integrate clinical features extracted from data acquired by different scanners and protocols to improve stability and robustness. Previous studies have described various computational approaches to fuse single modality multicentre datasets. However, these surveys rarely focused on evaluation metrics and lacked a checklist for computational data harmonisation studies. In this systematic review, we summarise the computational data harmonisation approaches for multi-modality data in the digital healthcare field, including harmonisation strategies and evaluation metrics based on different theories. In addition, a comprehensive checklist that summarises common practices for data harmonisation studies is proposed to guide researchers to report their research findings more effectively. Last but not least, flowcharts presenting possible ways for methodology and metric selection are proposed and the limitations of different methods have been surveyed for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge