Andreas Stefan
Optimization of Retrieval Algorithms on Large Scale Knowledge Graphs
Feb 10, 2020



Abstract:Knowledge graphs have been shown to play an important role in recent knowledge mining and discovery, for example in the field of life sciences or bioinformatics. Although a lot of research has been done on the field of query optimization, query transformation and of course in storing and retrieving large scale knowledge graphs the field of algorithmic optimization is still a major challenge and a vital factor in using graph databases. Few researchers have addressed the problem of optimizing algorithms on large scale labeled property graphs. Here, we present two optimization approaches and compare them with a naive approach of directly querying the graph database. The aim of our work is to determine limiting factors of graph databases like Neo4j and we describe a novel solution to tackle these challenges. For this, we suggest a classification schema to differ between the complexity of a problem on a graph database. We evaluate our optimization approaches on a test system containing a knowledge graph derived biomedical publication data enriched with text mining data. This dense graph has more than 71M nodes and 850M relationships. The results are very encouraging and - depending on the problem - we were able to show a speedup of a factor between 44 and 3839.
Data Exploration and Validation on dense knowledge graphs for biomedical research
Dec 08, 2019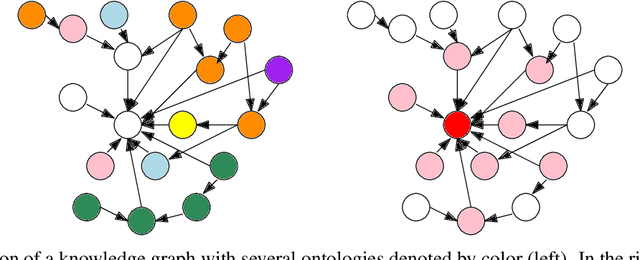
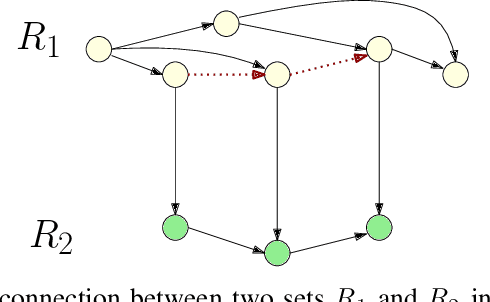
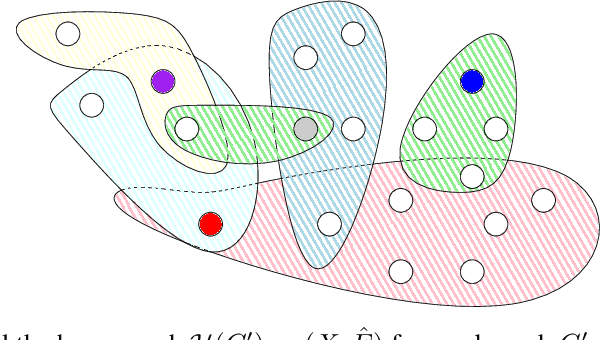
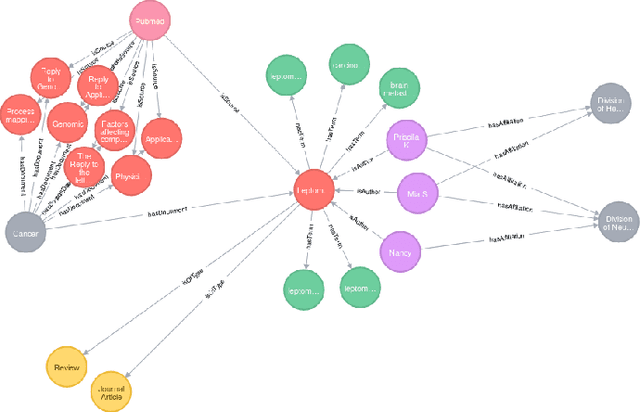
Abstract:Here we present a holistic approach for data exploration on dense knowledge graphs as a novel approach with a proof-of-concept in biomedical research. Knowledge graphs are increasingly becoming a vital factor in knowledge mining and discovery as they connect data using technologies from the semantic web. In this paper we extend a basic knowledge graph extracted from biomedical literature by context data like named entities and relations obtained by text mining and other linked data sources like ontologies and databases. We will present an overview about this novel network. The aim of this work was to extend this current knowledge with approaches from graph theory. This method will build the foundation for quality control, validation of hypothesis, detection of missing data and time series analysis of biomedical knowledge in general. In this context we tried to apply multiple-valued decision diagrams to these questions. In addition this knowledge representation of linked data can be used as FAIR approach to answer semantic questions. This paper sheds new lights on dense and very large knowledge graphs and the importance of a graph-theoretic understanding of these networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge