Amadou Barry
AdapDISCOM: An Adaptive Sparse Regression Method for High-Dimensional Multimodal Data With Block-Wise Missingness and Measurement Errors
Jul 31, 2025Abstract:Multimodal high-dimensional data are increasingly prevalent in biomedical research, yet they are often compromised by block-wise missingness and measurement errors, posing significant challenges for statistical inference and prediction. We propose AdapDISCOM, a novel adaptive direct sparse regression method that simultaneously addresses these two pervasive issues. Building on the DISCOM framework, AdapDISCOM introduces modality-specific weighting schemes to account for heterogeneity in data structures and error magnitudes across modalities. We establish the theoretical properties of AdapDISCOM, including model selection consistency and convergence rates under sub-Gaussian and heavy-tailed settings, and develop robust and computationally efficient variants (AdapDISCOM-Huber and Fast-AdapDISCOM). Extensive simulations demonstrate that AdapDISCOM consistently outperforms existing methods such as DISCOM, SCOM, and CoCoLasso, particularly under heterogeneous contamination and heavy-tailed distributions. Finally, we apply AdapDISCOM to Alzheimers Disease Neuroimaging Initiative (ADNI) data, demonstrating improved prediction of cognitive scores and reliable selection of established biomarkers, even with substantial missingness and measurement errors. AdapDISCOM provides a flexible, robust, and scalable framework for high-dimensional multimodal data analysis under realistic data imperfections.
An algorithm-based multiple detection influence measure for high dimensional regression using expectile
May 26, 2021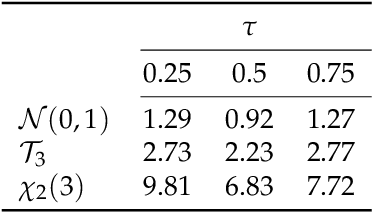
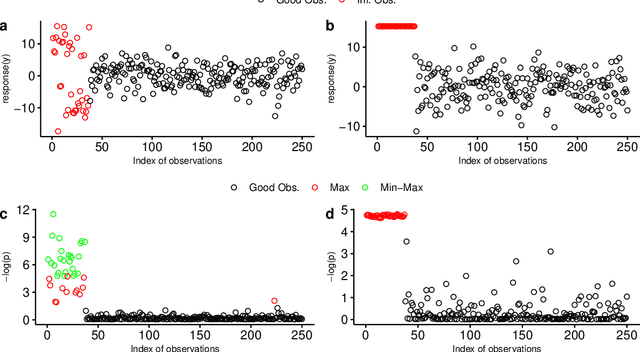
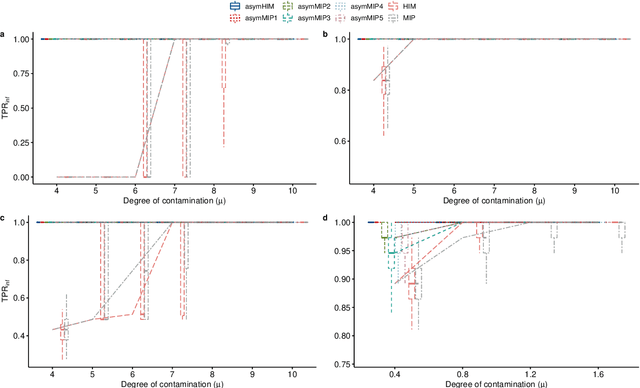
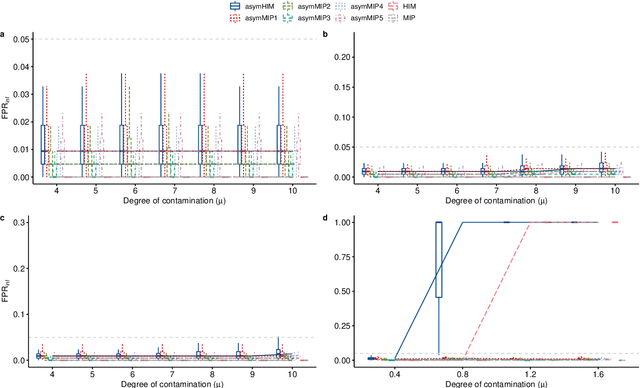
Abstract:The identification of influential observations is an important part of data analysis that can prevent erroneous conclusions drawn from biased estimators. However, in high dimensional data, this identification is challenging. Classical and recently-developed methods often perform poorly when there are multiple influential observations in the same dataset. In particular, current methods can fail when there is masking several influential observations with similar characteristics, or swamping when the influential observations are near the boundary of the space spanned by well-behaved observations. Therefore, we propose an algorithm-based, multi-step, multiple detection procedure to identify influential observations that addresses current limitations. Our three-step algorithm to identify and capture undesirable variability in the data, $\asymMIP,$ is based on two complementary statistics, inspired by asymmetric correlations, and built on expectiles. Simulations demonstrate higher detection power than competing methods. Use of the resulting asymptotic distribution leads to detection of influential observations without the need for computationally demanding procedures such as the bootstrap. The application of our method to the Autism Brain Imaging Data Exchange neuroimaging dataset resulted in a more balanced and accurate prediction of brain maturity based on cortical thickness. See our GitHub for a free R package that implements our algorithm: \texttt{asymMIP} (\url{github.com/AmBarry/hidetify}).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge