Alexandros Ioannidis
An Analysis of Indexing and Querying Strategies on a Technologically Assisted Review Task
Apr 20, 2021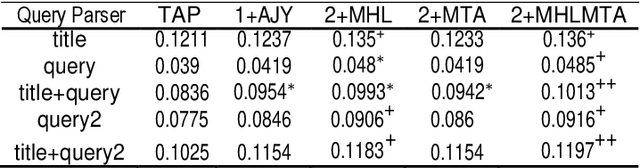
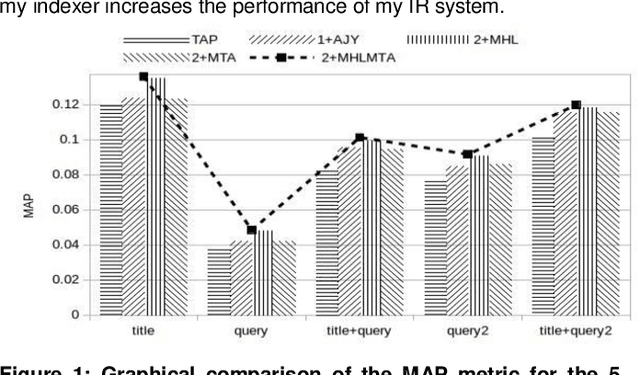
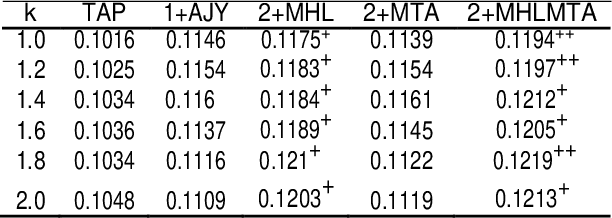
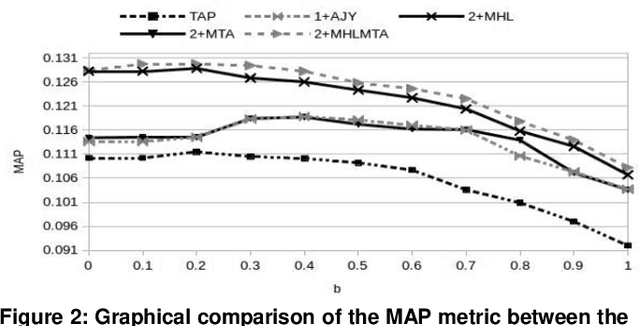
Abstract:This paper presents a preliminary experimentation study using the CLEF 2017 eHealth Task 2 collection for evaluating the effectiveness of different indexing methodologies of documents and query parsing techniques. Furthermore, it is an attempt to advance and share the efforts of observing the characteristics and helpfulness of various methodologies for indexing PubMed documents and for different topic parsing techniques to produce queries. For this purpose, my research includes experimentation with different document indexing methodologies, by utilising existing tools, such as the Lucene4IR (L4IR) information retrieval system, the Technology Assisted Reviews for Empirical Medicine tool for parsing topics of the CLEF collection and the TREC evaluation tool to appraise system's performance. The results showed that including a greater number of fields to the PubMed indexer of L4IR is a decisive factor for the retrieval effectiveness of L4IR.
An Analysis of a BERT Deep Learning Strategy on a Technology Assisted Review Task
Apr 16, 2021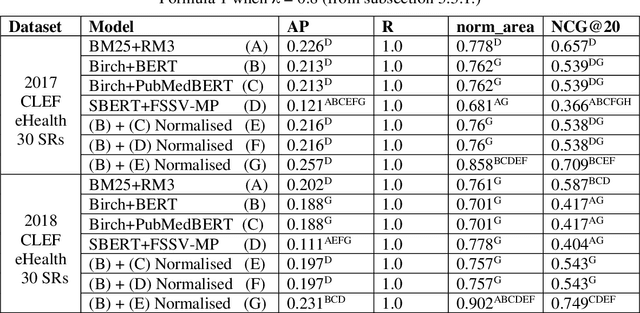
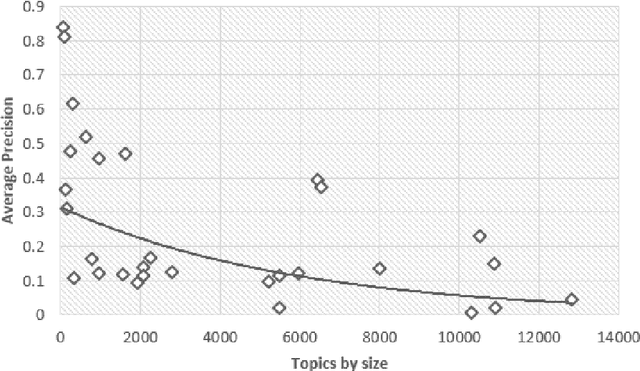
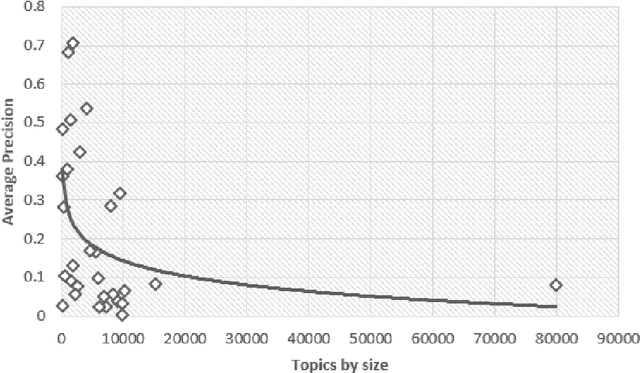
Abstract:Document screening is a central task within Evidenced Based Medicine, which is a clinical discipline that supplements scientific proof to back medical decisions. Given the recent advances in DL (Deep Learning) methods applied to Information Retrieval tasks, I propose a DL document classification approach with BERT or PubMedBERT embeddings and a DL similarity search path using SBERT embeddings to reduce physicians' tasks of screening and classifying immense amounts of documents to answer clinical queries. I test and evaluate the retrieval effectiveness of my DL strategy on the 2017 and 2018 CLEF eHealth collections. I find that the proposed DL strategy works, I compare it to the recently successful BM25 plus RM3 model, and conclude that the suggested method accomplishes advanced retrieval performance in the initial ranking of the articles with the aforementioned datasets, for the CLEF eHealth Technologically Assisted Reviews in Empirical Medicine Task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge