Alessandro Balestrucci
Agentomics-ML: Autonomous Machine Learning Experimentation Agent for Genomic and Transcriptomic Data
Jun 05, 2025Abstract:The adoption of machine learning (ML) and deep learning methods has revolutionized molecular medicine by driving breakthroughs in genomics, transcriptomics, drug discovery, and biological systems modeling. The increasing quantity, multimodality, and heterogeneity of biological datasets demand automated methods that can produce generalizable predictive models. Recent developments in large language model-based agents have shown promise for automating end-to-end ML experimentation on structured benchmarks. However, when applied to heterogeneous computational biology datasets, these methods struggle with generalization and success rates. Here, we introduce Agentomics-ML, a fully autonomous agent-based system designed to produce a classification model and the necessary files for reproducible training and inference. Our method follows predefined steps of an ML experimentation process, repeatedly interacting with the file system through Bash to complete individual steps. Once an ML model is produced, training and validation metrics provide scalar feedback to a reflection step to identify issues such as overfitting. This step then creates verbal feedback for future iterations, suggesting adjustments to steps such as data representation, model architecture, and hyperparameter choices. We have evaluated Agentomics-ML on several established genomic and transcriptomic benchmark datasets and show that it outperforms existing state-of-the-art agent-based methods in both generalization and success rates. While state-of-the-art models built by domain experts still lead in absolute performance on the majority of the computational biology datasets used in this work, Agentomics-ML narrows the gap for fully autonomous systems and achieves state-of-the-art performance on one of the used benchmark datasets. The code is available at https://github.com/BioGeMT/Agentomics-ML.
On the Robustness of Ensemble-Based Machine Learning Against Data Poisoning
Sep 28, 2022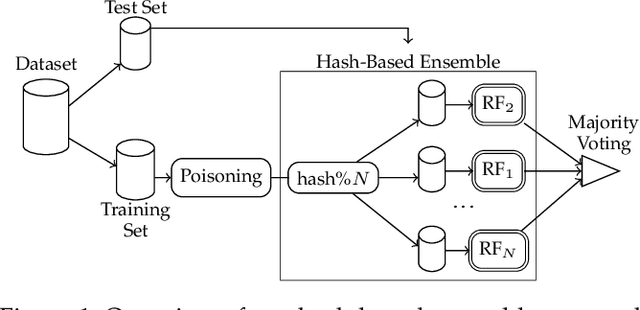
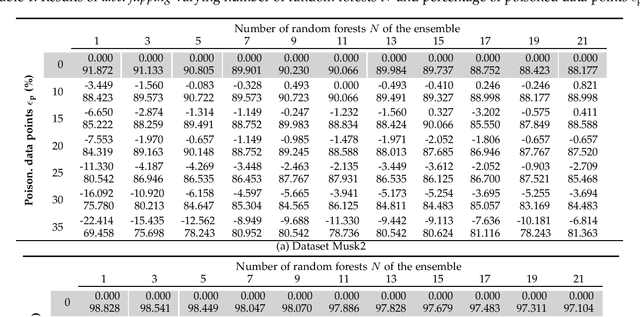
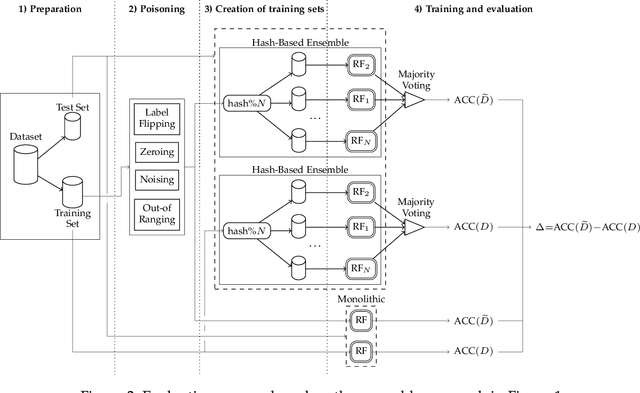
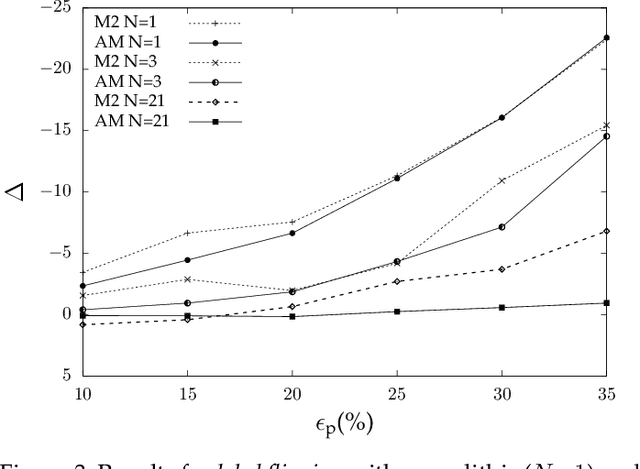
Abstract:Machine learning is becoming ubiquitous. From financial to medicine, machine learning models are boosting decision-making processes and even outperforming humans in some tasks. This huge progress in terms of prediction quality does not however find a counterpart in the security of such models and corresponding predictions, where perturbations of fractions of the training set (poisoning) can seriously undermine the model accuracy. Research on poisoning attacks and defenses even predates the introduction of deep neural networks, leading to several promising solutions. Among them, ensemble-based defenses, where different models are trained on portions of the training set and their predictions are then aggregated, are getting significant attention, due to their relative simplicity and theoretical and practical guarantees. The work in this paper designs and implements a hash-based ensemble approach for ML robustness and evaluates its applicability and performance on random forests, a machine learning model proved to be more resistant to poisoning attempts on tabular datasets. An extensive experimental evaluation is carried out to evaluate the robustness of our approach against a variety of attacks, and compare it with a traditional monolithic model based on random forests.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge