Aaron J. Masino
PhenoGnet: A Graph-Based Contrastive Learning Framework for Disease Similarity Prediction
Sep 17, 2025Abstract:Understanding disease similarity is critical for advancing diagnostics, drug discovery, and personalized treatment strategies. We present PhenoGnet, a novel graph-based contrastive learning framework designed to predict disease similarity by integrating gene functional interaction networks with the Human Phenotype Ontology (HPO). PhenoGnet comprises two key components: an intra-view model that separately encodes gene and phenotype graphs using Graph Convolutional Networks (GCNs) and Graph Attention Networks (GATs), and a cross view model implemented as a shared weight multilayer perceptron (MLP) that aligns gene and phenotype embeddings through contrastive learning. The model is trained using known gene phenotype associations as positive pairs and randomly sampled unrelated pairs as negatives. Diseases are represented by the mean embeddings of their associated genes and/or phenotypes, and pairwise similarity is computed via cosine similarity. Evaluation on a curated benchmark of 1,100 similar and 866 dissimilar disease pairs demonstrates strong performance, with gene based embeddings achieving an AUCPR of 0.9012 and AUROC of 0.8764, outperforming existing state of the art methods. Notably, PhenoGnet captures latent biological relationships beyond direct overlap, offering a scalable and interpretable solution for disease similarity prediction. These results underscore its potential for enabling downstream applications in rare disease research and precision medicine.
Deployment of a Robust and Explainable Mortality Prediction Model: The COVID-19 Pandemic and Beyond
Nov 28, 2023Abstract:This study investigated the performance, explainability, and robustness of deployed artificial intelligence (AI) models in predicting mortality during the COVID-19 pandemic and beyond. The first study of its kind, we found that Bayesian Neural Networks (BNNs) and intelligent training techniques allowed our models to maintain performance amidst significant data shifts. Our results emphasize the importance of developing robust AI models capable of matching or surpassing clinician predictions, even under challenging conditions. Our exploration of model explainability revealed that stochastic models generate more diverse and personalized explanations thereby highlighting the need for AI models that provide detailed and individualized insights in real-world clinical settings. Furthermore, we underscored the importance of quantifying uncertainty in AI models which enables clinicians to make better-informed decisions based on reliable predictions. Our study advocates for prioritizing implementation science in AI research for healthcare and ensuring that AI solutions are practical, beneficial, and sustainable in real-world clinical environments. By addressing unique challenges and complexities in healthcare settings, researchers can develop AI models that effectively improve clinical practice and patient outcomes.
Revisiting the Fragility of Influence Functions
Mar 22, 2023



Abstract:In the last few years, many works have tried to explain the predictions of deep learning models. Few methods, however, have been proposed to verify the accuracy or faithfulness of these explanations. Recently, influence functions, which is a method that approximates the effect that leave-one-out training has on the loss function, has been shown to be fragile. The proposed reason for their fragility remains unclear. Although previous work suggests the use of regularization to increase robustness, this does not hold in all cases. In this work, we seek to investigate the experiments performed in the prior work in an effort to understand the underlying mechanisms of influence function fragility. First, we verify influence functions using procedures from the literature under conditions where the convexity assumptions of influence functions are met. Then, we relax these assumptions and study the effects of non-convexity by using deeper models and more complex datasets. Here, we analyze the key metrics and procedures that are used to validate influence functions. Our results indicate that the validation procedures may cause the observed fragility.
Unsupervised learning with GLRM feature selection reveals novel traumatic brain injury phenotypes
Nov 30, 2018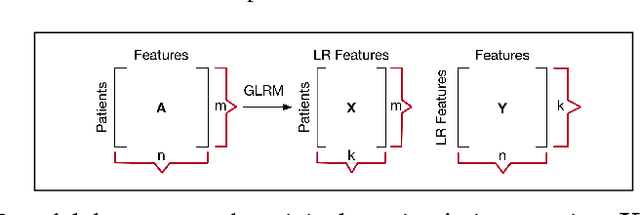
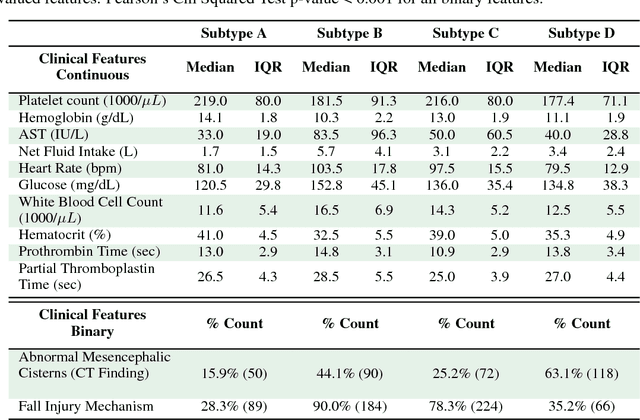
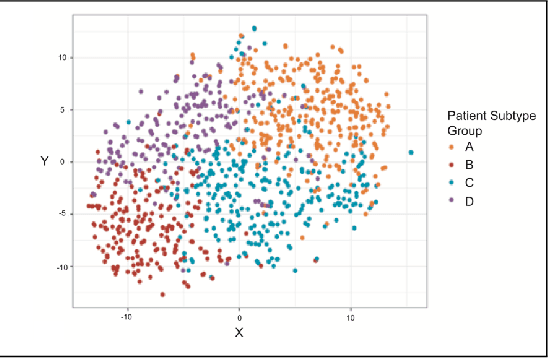
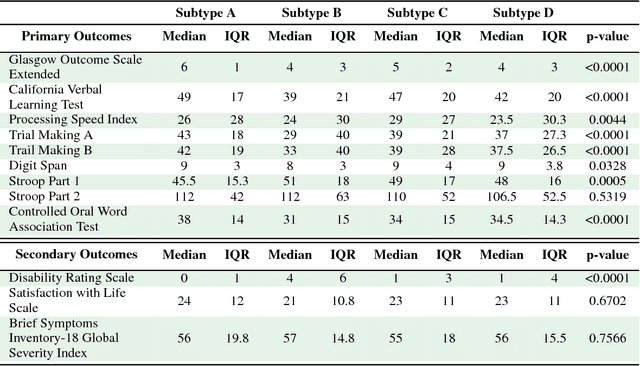
Abstract:Baseline injury categorization is important to traumatic brain injury (TBI) research and treatment. Current categorization is dominated by symptom-based scores that insufficiently capture injury heterogeneity. In this work, we apply unsupervised clustering to identify novel TBI phenotypes. Our approach uses a generalized low-rank model (GLRM) model for feature selection in a procedure analogous to wrapper methods. The resulting clusters reveal four novel TBI phenotypes with distinct feature profiles and that correlate to 90-day functional and cognitive status.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge