Bowen Jing
Verlet Flows: Exact-Likelihood Integrators for Flow-Based Generative Models
May 05, 2024Approximations in computing model likelihoods with continuous normalizing flows (CNFs) hinder the use of these models for importance sampling of Boltzmann distributions, where exact likelihoods are required. In this work, we present Verlet flows, a class of CNFs on an augmented state-space inspired by symplectic integrators from Hamiltonian dynamics. When used with carefully constructed Taylor-Verlet integrators, Verlet flows provide exact-likelihood generative models which generalize coupled flow architectures from a non-continuous setting while imposing minimal expressivity constraints. On experiments over toy densities, we demonstrate that the variance of the commonly used Hutchinson trace estimator is unsuitable for importance sampling, whereas Verlet flows perform comparably to full autograd trace computations while being significantly faster.
Dirichlet Flow Matching with Applications to DNA Sequence Design
Feb 08, 2024Discrete diffusion or flow models could enable faster and more controllable sequence generation than autoregressive models. We show that na\"ive linear flow matching on the simplex is insufficient toward this goal since it suffers from discontinuities in the training target and further pathologies. To overcome this, we develop Dirichlet flow matching on the simplex based on mixtures of Dirichlet distributions as probability paths. In this framework, we derive a connection between the mixtures' scores and the flow's vector field that allows for classifier and classifier-free guidance. Further, we provide distilled Dirichlet flow matching, which enables one-step sequence generation with minimal performance hits, resulting in $O(L)$ speedups compared to autoregressive models. On complex DNA sequence generation tasks, we demonstrate superior performance compared to all baselines in distributional metrics and in achieving desired design targets for generated sequences. Finally, we show that our classifier-free guidance approach improves unconditional generation and is effective for generating DNA that satisfies design targets. Code is available at https://github.com/HannesStark/dirichlet-flow-matching.
AlphaFold Meets Flow Matching for Generating Protein Ensembles
Feb 07, 2024The biological functions of proteins often depend on dynamic structural ensembles. In this work, we develop a flow-based generative modeling approach for learning and sampling the conformational landscapes of proteins. We repurpose highly accurate single-state predictors such as AlphaFold and ESMFold and fine-tune them under a custom flow matching framework to obtain sequence-conditoned generative models of protein structure called AlphaFlow and ESMFlow. When trained and evaluated on the PDB, our method provides a superior combination of precision and diversity compared to AlphaFold with MSA subsampling. When further trained on ensembles from all-atom MD, our method accurately captures conformational flexibility, positional distributions, and higher-order ensemble observables for unseen proteins. Moreover, our method can diversify a static PDB structure with faster wall-clock convergence to certain equilibrium properties than replicate MD trajectories, demonstrating its potential as a proxy for expensive physics-based simulations. Code is available at https://github.com/bjing2016/alphaflow.
Equivariant Scalar Fields for Molecular Docking with Fast Fourier Transforms
Dec 07, 2023Molecular docking is critical to structure-based virtual screening, yet the throughput of such workflows is limited by the expensive optimization of scoring functions involved in most docking algorithms. We explore how machine learning can accelerate this process by learning a scoring function with a functional form that allows for more rapid optimization. Specifically, we define the scoring function to be the cross-correlation of multi-channel ligand and protein scalar fields parameterized by equivariant graph neural networks, enabling rapid optimization over rigid-body degrees of freedom with fast Fourier transforms. The runtime of our approach can be amortized at several levels of abstraction, and is particularly favorable for virtual screening settings with a common binding pocket. We benchmark our scoring functions on two simplified docking-related tasks: decoy pose scoring and rigid conformer docking. Our method attains similar but faster performance on crystal structures compared to the widely-used Vina and Gnina scoring functions, and is more robust on computationally predicted structures. Code is available at https://github.com/bjing2016/scalar-fields.
Harmonic Self-Conditioned Flow Matching for Multi-Ligand Docking and Binding Site Design
Oct 09, 2023



A significant amount of protein function requires binding small molecules, including enzymatic catalysis. As such, designing binding pockets for small molecules has several impactful applications ranging from drug synthesis to energy storage. Towards this goal, we first develop HarmonicFlow, an improved generative process over 3D protein-ligand binding structures based on our self-conditioned flow matching objective. FlowSite extends this flow model to jointly generate a protein pocket's discrete residue types and the molecule's binding 3D structure. We show that HarmonicFlow improves upon the state-of-the-art generative processes for docking in simplicity, generality, and performance. Enabled by this structure modeling, FlowSite designs binding sites substantially better than baseline approaches and provides the first general solution for binding site design.
nnSAM: Plug-and-play Segment Anything Model Improves nnUNet Performance
Oct 02, 2023



The recent developments of foundation models in computer vision, especially the Segment Anything Model (SAM), allow scalable and domain-agnostic image segmentation to serve as a general-purpose segmentation tool. In parallel, the field of medical image segmentation has benefited significantly from specialized neural networks like the nnUNet, which is trained on domain-specific datasets and can automatically configure the network to tailor to specific segmentation challenges. To combine the advantages of foundation models and domain-specific models, we present nnSAM, which synergistically integrates the SAM model with the nnUNet model to achieve more accurate and robust medical image segmentation. The nnSAM model leverages the powerful and robust feature extraction capabilities of SAM, while harnessing the automatic configuration capabilities of nnUNet to promote dataset-tailored learning. Our comprehensive evaluation of nnSAM model on different sizes of training samples shows that it allows few-shot learning, which is highly relevant for medical image segmentation where high-quality, annotated data can be scarce and costly to obtain. By melding the strengths of both its predecessors, nnSAM positions itself as a potential new benchmark in medical image segmentation, offering a tool that combines broad applicability with specialized efficiency. The code is available at https://github.com/Kent0n-Li/Medical-Image-Segmentation.
EigenFold: Generative Protein Structure Prediction with Diffusion Models
Apr 05, 2023



Protein structure prediction has reached revolutionary levels of accuracy on single structures, yet distributional modeling paradigms are needed to capture the conformational ensembles and flexibility that underlie biological function. Towards this goal, we develop EigenFold, a diffusion generative modeling framework for sampling a distribution of structures from a given protein sequence. We define a diffusion process that models the structure as a system of harmonic oscillators and which naturally induces a cascading-resolution generative process along the eigenmodes of the system. On recent CAMEO targets, EigenFold achieves a median TMScore of 0.84, while providing a more comprehensive picture of model uncertainty via the ensemble of sampled structures relative to existing methods. We then assess EigenFold's ability to model and predict conformational heterogeneity for fold-switching proteins and ligand-induced conformational change. Code is available at https://github.com/bjing2016/EigenFold.
DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking
Oct 04, 2022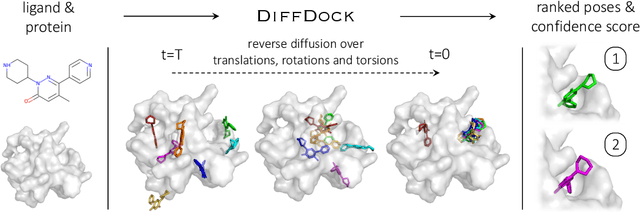
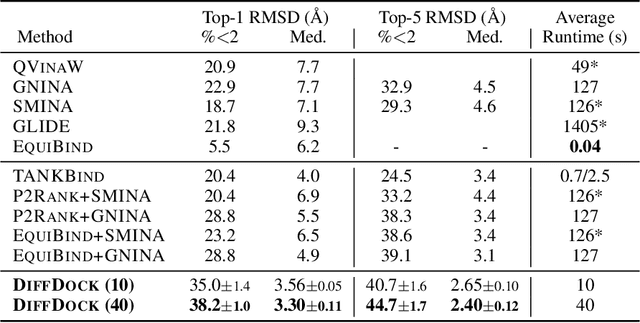
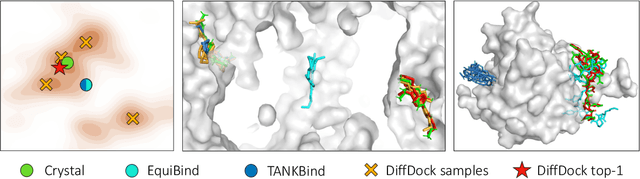
Predicting the binding structure of a small molecule ligand to a protein -- a task known as molecular docking -- is critical to drug design. Recent deep learning methods that treat docking as a regression problem have decreased runtime compared to traditional search-based methods but have yet to offer substantial improvements in accuracy. We instead frame molecular docking as a generative modeling problem and develop DiffDock, a diffusion generative model over the non-Euclidean manifold of ligand poses. To do so, we map this manifold to the product space of the degrees of freedom (translational, rotational, and torsional) involved in docking and develop an efficient diffusion process on this space. Empirically, DiffDock obtains a 38% top-1 success rate (RMSD<2A) on PDBBind, significantly outperforming the previous state-of-the-art of traditional docking (23%) and deep learning (20%) methods. Moreover, DiffDock has fast inference times and provides confidence estimates with high selective accuracy.
Torsional Diffusion for Molecular Conformer Generation
Jun 01, 2022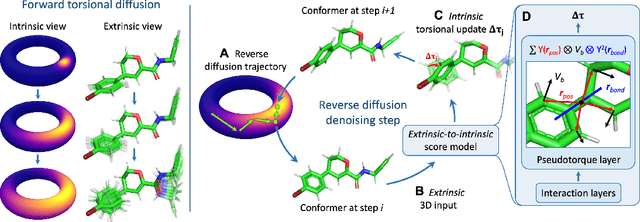

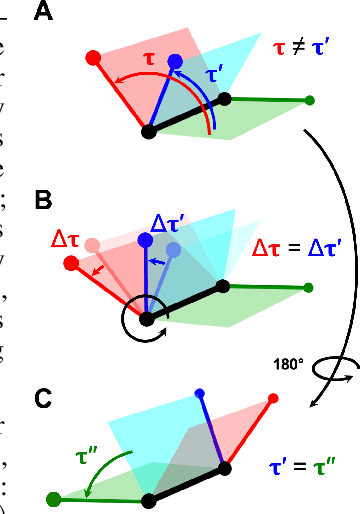
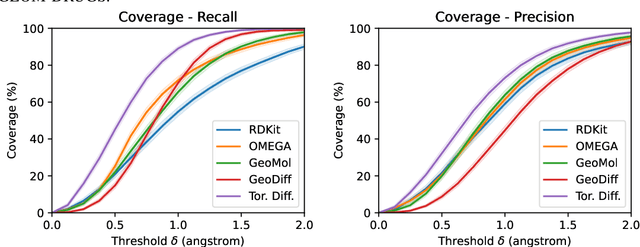
Molecular conformer generation is a fundamental task in computational chemistry. Several machine learning approaches have been developed, but none have outperformed state-of-the-art cheminformatics methods. We propose torsional diffusion, a novel diffusion framework that operates on the space of torsion angles via a diffusion process on the hypertorus and an extrinsic-to-intrinsic score model. On a standard benchmark of drug-like molecules, torsional diffusion generates superior conformer ensembles compared to machine learning and cheminformatics methods in terms of both RMSD and chemical properties, and is orders of magnitude faster than previous diffusion-based models. Moreover, our model provides exact likelihoods, which we employ to build the first generalizable Boltzmann generator. Code is available at https://github.com/gcorso/torsional-diffusion.
Subspace Diffusion Generative Models
May 03, 2022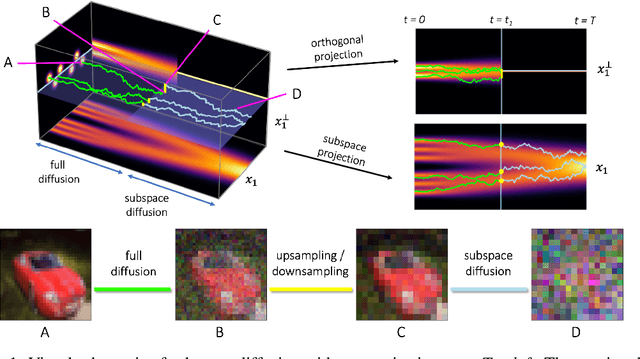
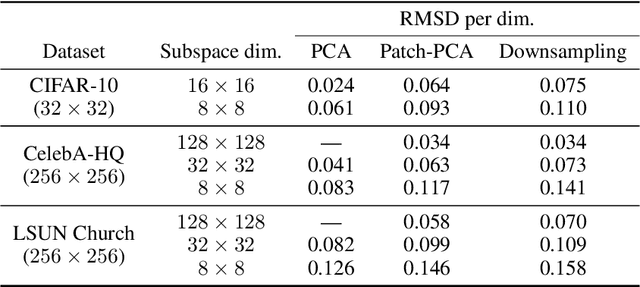
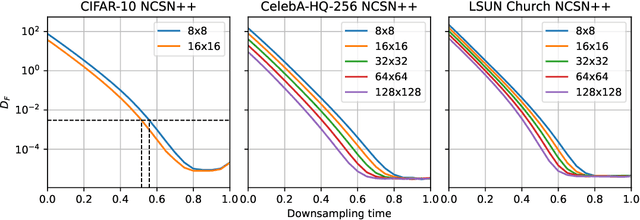
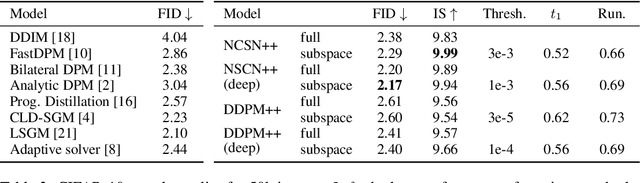
Score-based models generate samples by mapping noise to data (and vice versa) via a high-dimensional diffusion process. We question whether it is necessary to run this entire process at high dimensionality and incur all the inconveniences thereof. Instead, we restrict the diffusion via projections onto subspaces as the data distribution evolves toward noise. When applied to state-of-the-art models, our framework simultaneously improves sample quality -- reaching an FID of 2.17 on unconditional CIFAR-10 -- and reduces the computational cost of inference for the same number of denoising steps. Our framework is fully compatible with continuous-time diffusion and retains its flexible capabilities, including exact log-likelihoods and controllable generation. Code is available at https://github.com/bjing2016/subspace-diffusion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge