"cancer detection": models, code, and papers
Distill-to-Label: Weakly Supervised Instance Labeling Using Knowledge Distillation
Jul 26, 2019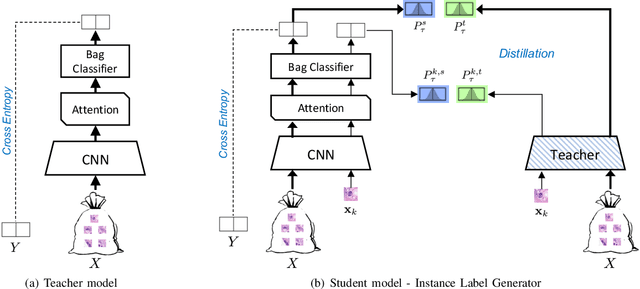
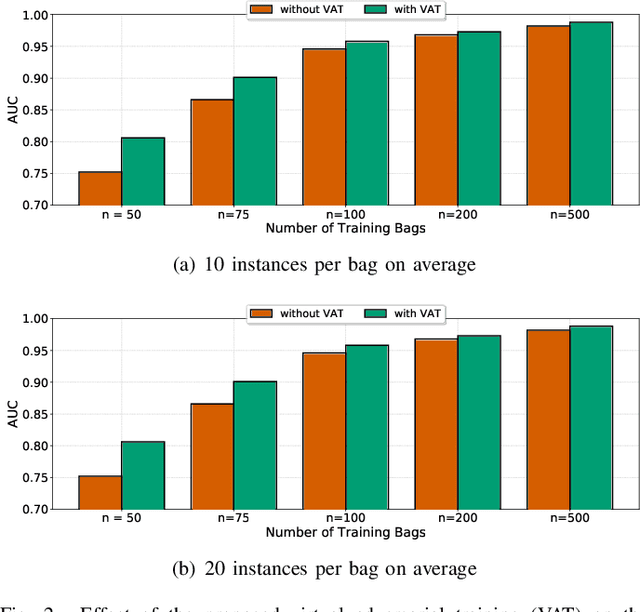
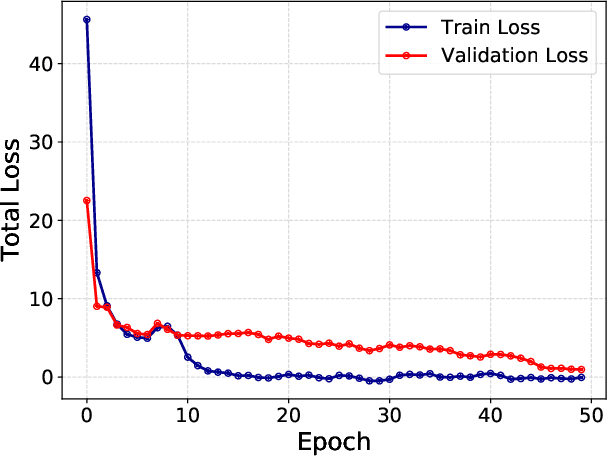
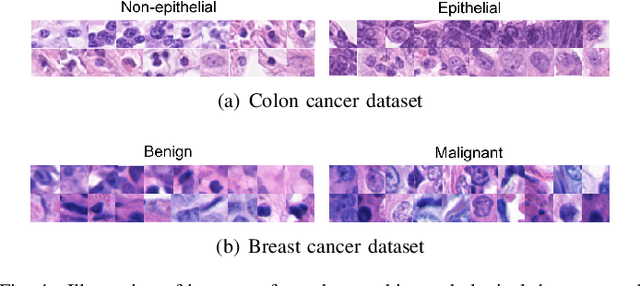
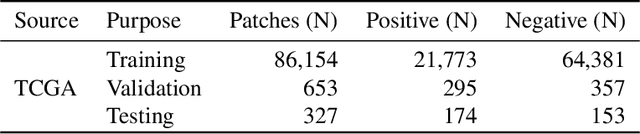
Weakly supervised instance labeling using only image-level labels, in lieu of expensive fine-grained pixel annotations, is crucial in several applications including medical image analysis. In contrast to conventional instance segmentation scenarios in computer vision, the problems that we consider are characterized by a small number of training images and non-local patterns that lead to the diagnosis. In this paper, we explore the use of multiple instance learning (MIL) to design an instance label generator under this weakly supervised setting. Motivated by the observation that an MIL model can handle bags of varying sizes, we propose to repurpose an MIL model originally trained for bag-level classification to produce reliable predictions for single instances, i.e., bags of size $1$. To this end, we introduce a novel regularization strategy based on virtual adversarial training for improving MIL training, and subsequently develop a knowledge distillation technique for repurposing the trained MIL model. Using empirical studies on colon cancer and breast cancer detection from histopathological images, we show that the proposed approach produces high-quality instance-level prediction and significantly outperforms state-of-the MIL methods.
Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging
Sep 27, 2019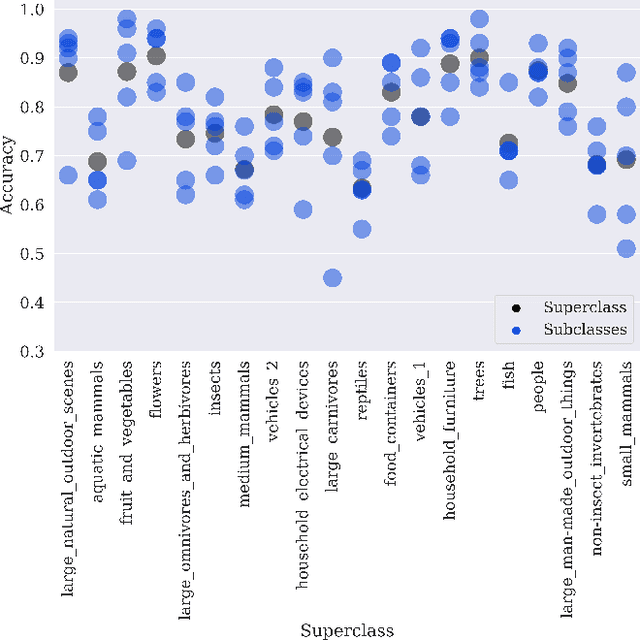
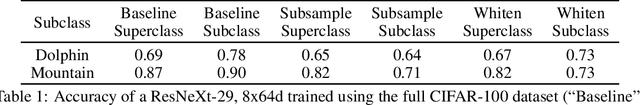
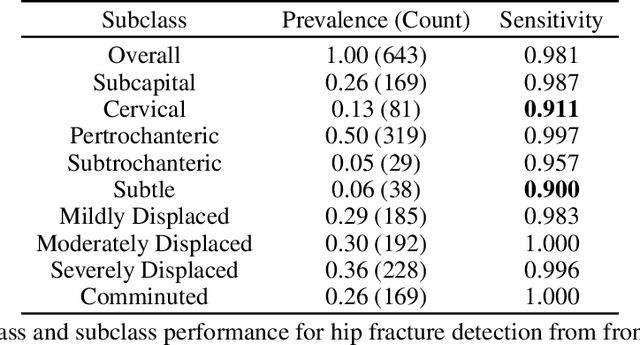
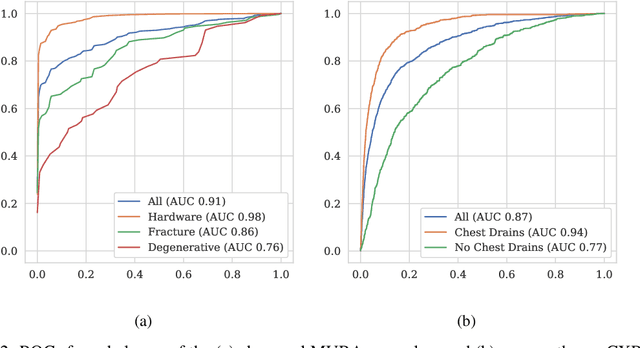
Machine learning models for medical image analysis often suffer from poor performance on important subsets of a population that are not identified during training or testing. For example, overall performance of a cancer detection model may be high, but the model still consistently misses a rare but aggressive cancer subtype. We refer to this problem as hidden stratification, and observe that it results from incompletely describing the meaningful variation in a dataset. While hidden stratification can substantially reduce the clinical efficacy of machine learning models, its effects remain difficult to measure. In this work, we assess the utility of several possible techniques for measuring and describing hidden stratification effects, and characterize these effects both on multiple medical imaging datasets and via synthetic experiments on the well-characterised CIFAR-100 benchmark dataset. We find evidence that hidden stratification can occur in unidentified imaging subsets with low prevalence, low label quality, subtle distinguishing features, or spurious correlates, and that it can result in relative performance differences of over 20% on clinically important subsets. Finally, we explore the clinical implications of our findings, and suggest that evaluation of hidden stratification should be a critical component of any machine learning deployment in medical imaging.
Perineural Invasion Detection in Multiple Organ Cancer Based on Deep Convolutional Neural Network
Oct 23, 2021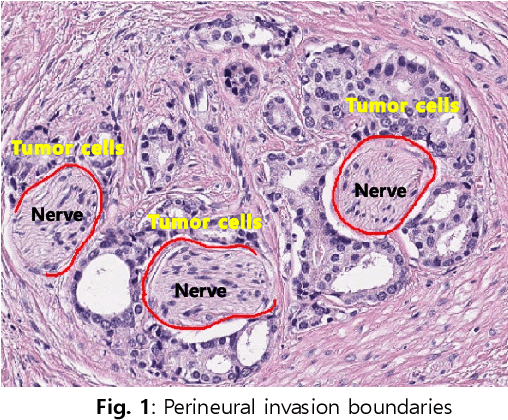

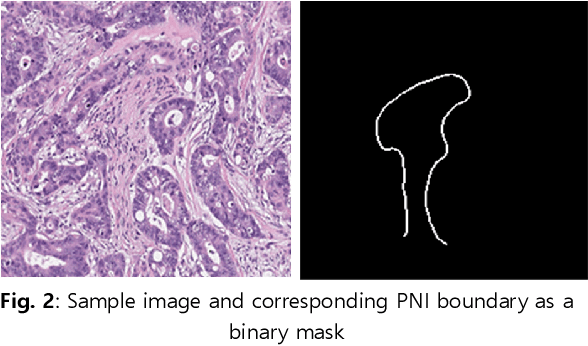
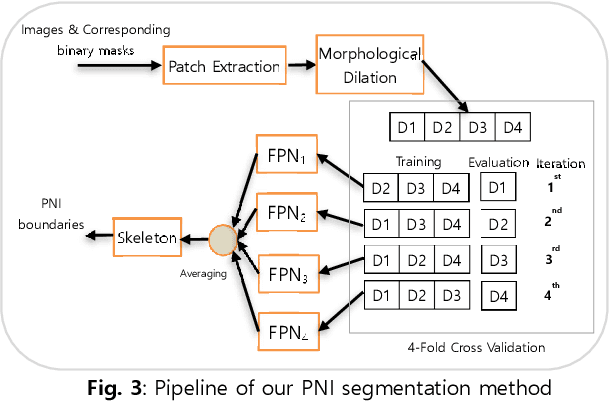
Perineural invasion (PNI) by malignant tumor cells has been reported as an independent indicator of poor prognosis in various cancers. Assessment of PNI in small nerves on glass slides is a labor-intensive task. In this study, we propose an algorithm to detect the perineural invasions in colon, prostate, and pancreas cancers based on a convolutional neural network (CNN).
Early Diagnosis of Lung Cancer Using Computer Aided Detection via Lung Segmentation Approach
Jul 23, 2021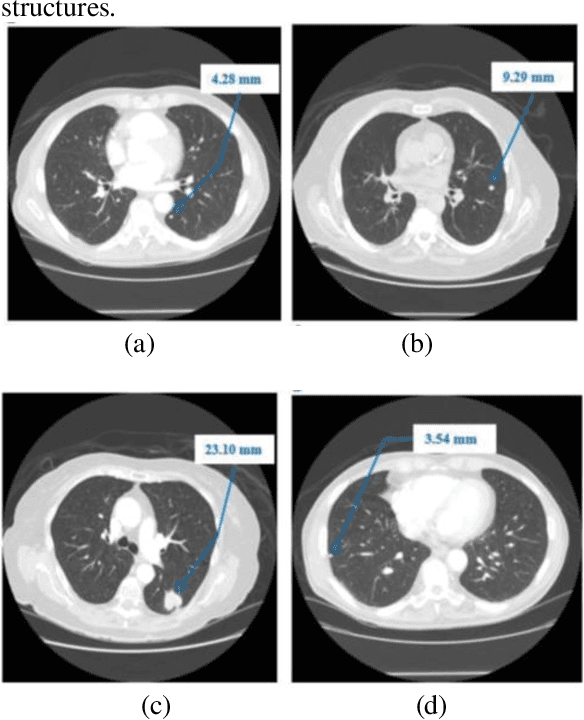
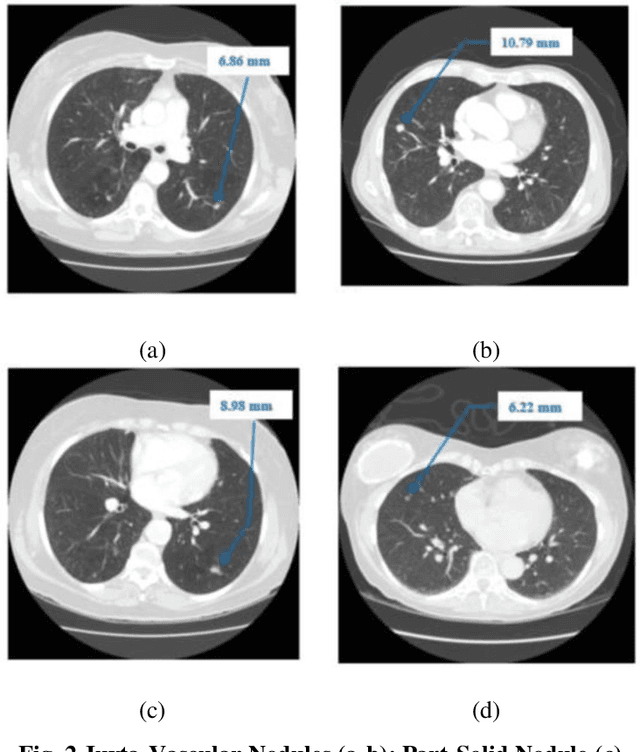
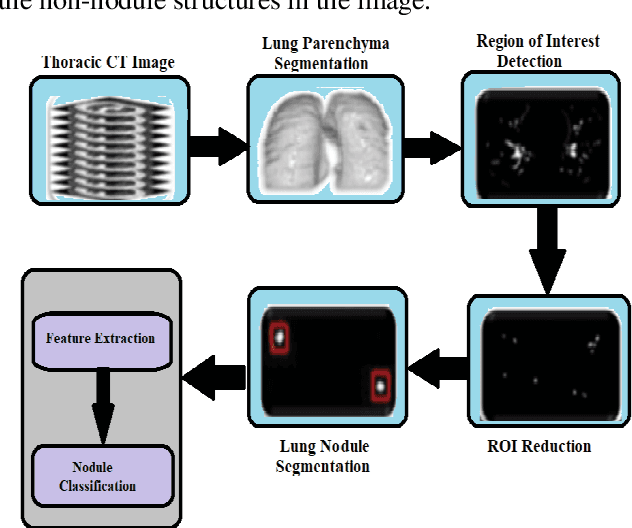
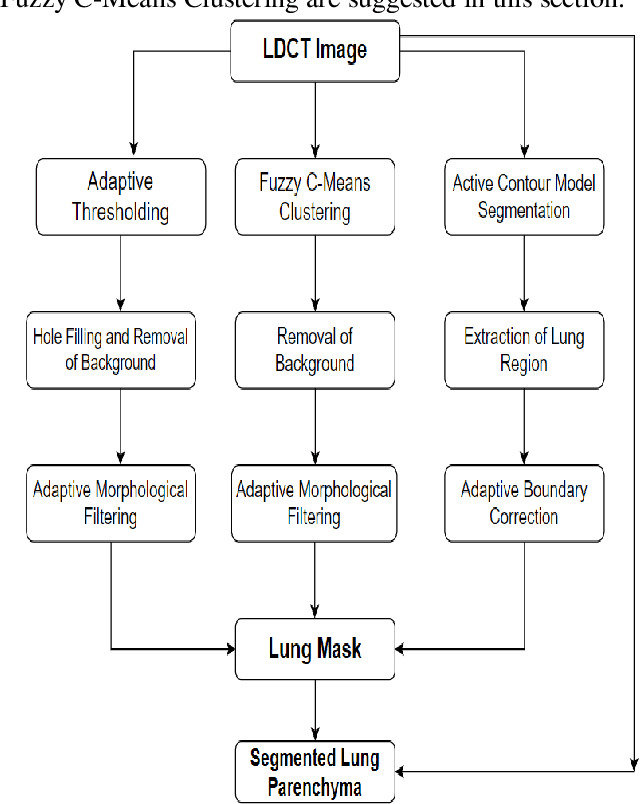
Lung cancer begins in the lungs and leading to the reason of cancer demise amid population in the creation. According to the American Cancer Society, which estimates about 27% of the deaths because of cancer. In the early phase of its evolution, lung cancer does not cause any symptoms usually. Many of the patients have been diagnosed in a developed phase where symptoms become more prominent, that results in poor curative treatment and high mortality rate. Computer Aided Detection systems are used to achieve greater accuracies for the lung cancer diagnosis. In this research exertion, we proposed a novel methodology for lung Segmentation on the basis of Fuzzy C-Means Clustering, Adaptive Thresholding, and Segmentation of Active Contour Model. The experimental results are analysed and presented.
* 9 pages, 10 figures, Published with International Journal of Engineering Trends and Technology (IJETT)
Utilizing Automated Breast Cancer Detection to Identify Spatial Distributions of Tumor Infiltrating Lymphocytes in Invasive Breast Cancer
May 29, 2019
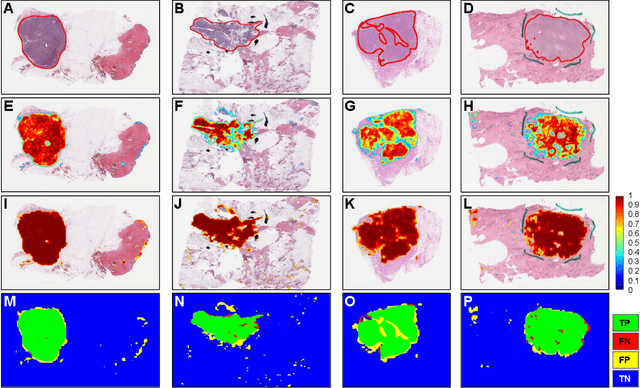
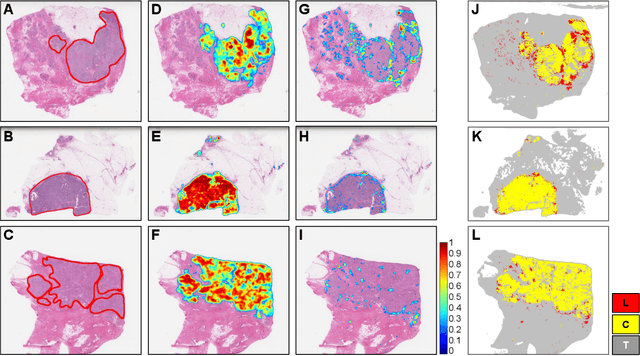
Quantitative assessment of Tumor-TIL spatial relationships is increasingly important in both basic science and clinical aspects of breast cancer research. We have developed and evaluated convolutional neural network (CNN) analysis pipelines to generate combined maps of cancer regions and tumor infiltrating lymphocytes (TILs) in routine diagnostic breast cancer whole slide tissue images (WSIs). We produce interactive whole slide maps that provide 1) insight about the structural patterns and spatial distribution of lymphocytic infiltrates and 2) facilitate improved quantification of TILs. We evaluated both tumor and TIL analyses using three CNN networks - Resnet-34, VGG16 and Inception v4, and demonstrated that the results compared favorably to those obtained by what believe are the best published methods. We have produced open-source tools and generated a public dataset consisting of tumor/TIL maps for 1,015 TCGA breast cancer images. We also present a customized web-based interface that enables easy visualization and interactive exploration of high-resolution combined Tumor-TIL maps for 1,015TCGA invasive breast cancer cases that can be downloaded for further downstream analyses.
Multiple Myeloma Cancer Cell Instance Segmentation
Sep 19, 2021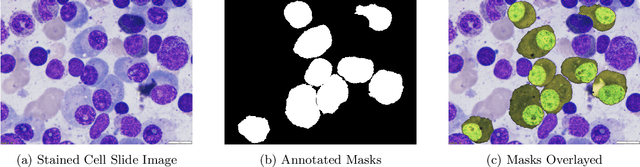
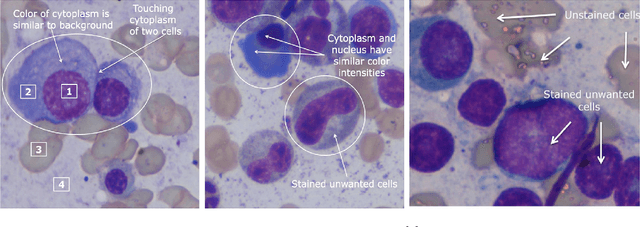
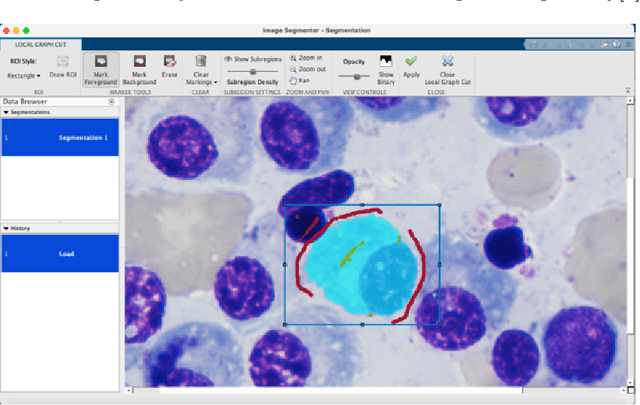

Images remain the largest data source in the field of healthcare. But at the same time, they are the most difficult to analyze. More than often, these images are analyzed by human experts such as pathologists and physicians. But due to considerable variation in pathology and the potential fatigue of human experts, an automated solution is much needed. The recent advancement in Deep learning could help us achieve an efficient and economical solution for the same. In this research project, we focus on developing a Deep Learning-based solution for detecting Multiple Myeloma cancer cells using an Object Detection and Instance Segmentation System. We explore multiple existing solutions and architectures for the task of Object Detection and Instance Segmentation and try to leverage them and come up with a novel architecture to achieve comparable and competitive performance on the required task. To train our model to detect and segment Multiple Myeloma cancer cells, we utilize a dataset curated by us using microscopic images of cell slides provided by Dr.Ritu Gupta(Prof., Dept. of Oncology AIIMS).
Whole-Sample Mapping of Cancerous and Benign Tissue Properties
Jul 23, 2019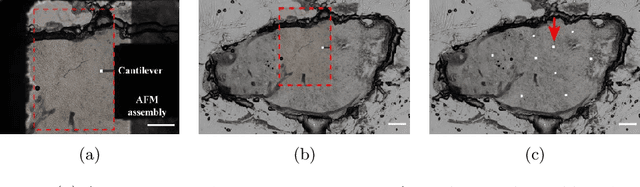
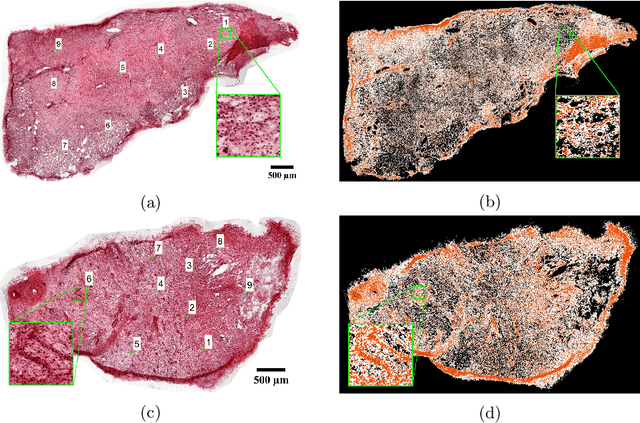
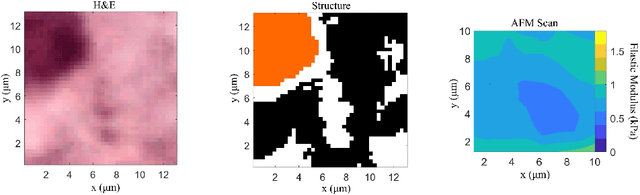

Structural and mechanical differences between cancerous and healthy tissue give rise to variations in macroscopic properties such as visual appearance and elastic modulus that show promise as signatures for early cancer detection. Atomic force microscopy (AFM) has been used to measure significant differences in stiffness between cancerous and healthy cells owing to its high force sensitivity and spatial resolution, however due to absorption and scattering of light, it is often challenging to accurately locate where AFM measurements have been made on a bulk tissue sample. In this paper we describe an image registration method that localizes AFM elastic stiffness measurements with high-resolution images of haematoxylin and eosin (H\&E)-stained tissue to within 1.5 microns. Color RGB images are segmented into three structure types (lumen, cells and stroma) by a neural network classifier trained on ground-truth pixel data obtained through k-means clustering in HSV color space. Using the localized stiffness maps and corresponding structural information, a whole-sample stiffness map is generated with a region matching and interpolation algorithm that associates similar structures with measured stiffness values. We present results showing significant differences in stiffness between healthy and cancerous liver tissue and discuss potential applications of this technique.
Automated Skin Lesion Classification Using Ensemble of Deep Neural Networks in ISIC 2018: Skin Lesion Analysis Towards Melanoma Detection Challenge
Jan 30, 2019
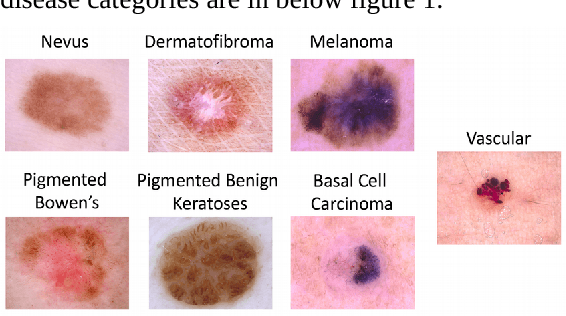
In this paper, we studied extensively on different deep learning based methods to detect melanoma and skin lesion cancers. Melanoma, a form of malignant skin cancer is very threatening to health. Proper diagnosis of melanoma at an earlier stage is crucial for the success rate of complete cure. Dermoscopic images with Benign and malignant forms of skin cancer can be analyzed by computer vision system to streamline the process of skin cancer detection. In this study, we experimented with various neural networks which employ recent deep learning based models like PNASNet-5-Large, InceptionResNetV2, SENet154, InceptionV4. Dermoscopic images are properly processed and augmented before feeding them into the network. We tested our methods on International Skin Imaging Collaboration (ISIC) 2018 challenge dataset. Our system has achieved best validation score of 0.76 for PNASNet-5-Large model. Further improvement and optimization of the proposed methods with a bigger training dataset and carefully chosen hyper-parameter could improve the performances. The code available for download at https://github.com/miltonbd/ISIC_2018_classification
A Hierarchical Multi-Task Approach to Gastrointestinal Image Analysis
Nov 16, 2021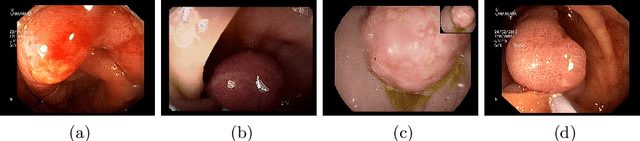
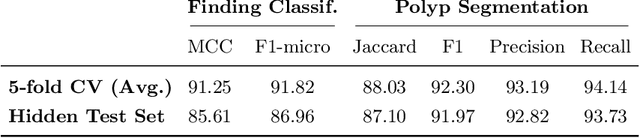
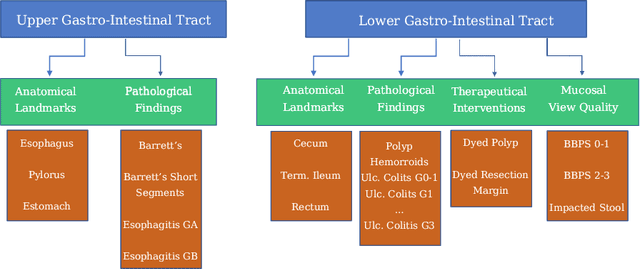
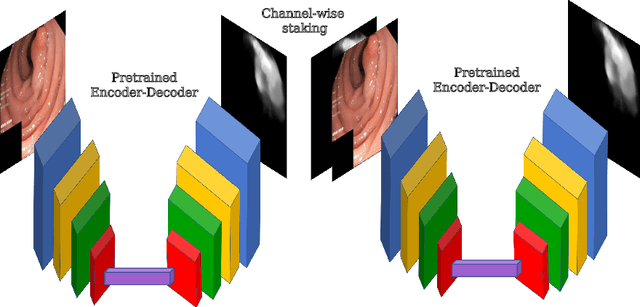
A large number of different lesions and pathologies can affect the human digestive system, resulting in life-threatening situations. Early detection plays a relevant role in the successful treatment and the increase of current survival rates to, e.g., colorectal cancer. The standard procedure enabling detection, endoscopic video analysis, generates large quantities of visual data that need to be carefully analyzed by an specialist. Due to the wide range of color, shape, and general visual appearance of pathologies, as well as highly varying image quality, such process is greatly dependent on the human operator experience and skill. In this work, we detail our solution to the task of multi-category classification of images from the gastrointestinal (GI) human tract within the 2020 Endotect Challenge. Our approach is based on a Convolutional Neural Network minimizing a hierarchical error function that takes into account not only the finding category, but also its location within the GI tract (lower/upper tract), and the type of finding (pathological finding/therapeutic intervention/anatomical landmark/mucosal views' quality). We also describe in this paper our solution for the challenge task of polyp segmentation in colonoscopies, which was addressed with a pretrained double encoder-decoder network. Our internal cross-validation results show an average performance of 91.25 Mathews Correlation Coefficient (MCC) and 91.82 Micro-F1 score for the classification task, and a 92.30 F1 score for the polyp segmentation task. The organization provided feedback on the performance in a hidden test set for both tasks, which resulted in 85.61 MCC and 86.96 F1 score for classification, and 91.97 F1 score for polyp segmentation. At the time of writing no public ranking for this challenge had been released.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge