"cancer detection": models, code, and papers
A Multi-resolution Model for Histopathology Image Classification and Localization with Multiple Instance Learning
Nov 05, 2020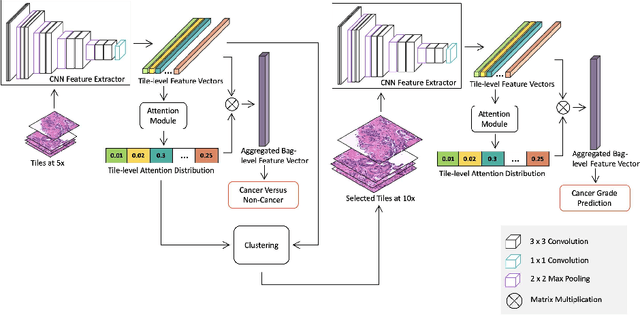
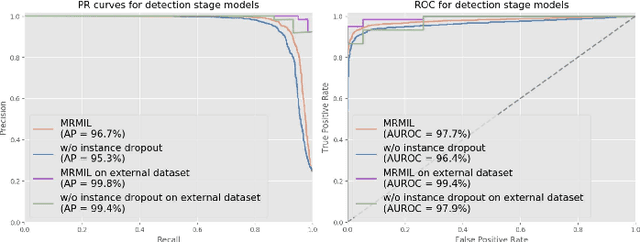
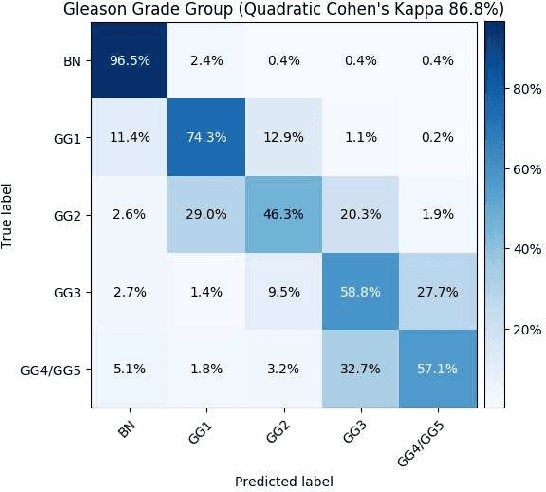
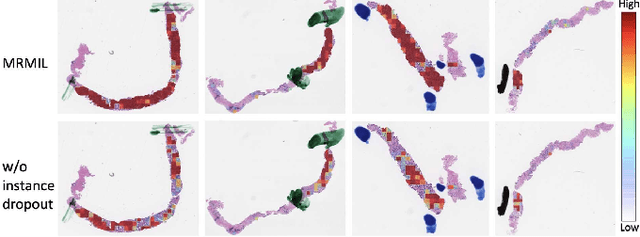
Histopathological images provide rich information for disease diagnosis. Large numbers of histopathological images have been digitized into high resolution whole slide images, opening opportunities in developing computational image analysis tools to reduce pathologists' workload and potentially improve inter- and intra- observer agreement. Most previous work on whole slide image analysis has focused on classification or segmentation of small pre-selected regions-of-interest, which requires fine-grained annotation and is non-trivial to extend for large-scale whole slide analysis. In this paper, we proposed a multi-resolution multiple instance learning model that leverages saliency maps to detect suspicious regions for fine-grained grade prediction. Instead of relying on expensive region- or pixel-level annotations, our model can be trained end-to-end with only slide-level labels. The model is developed on a large-scale prostate biopsy dataset containing 20,229 slides from 830 patients. The model achieved 92.7% accuracy, 81.8% Cohen's Kappa for benign, low grade (i.e. Grade group 1) and high grade (i.e. Grade group >= 2) prediction, an area under the receiver operating characteristic curve (AUROC) of 98.2% and an average precision (AP) of 97.4% for differentiating malignant and benign slides. The model obtained an AUROC of 99.4% and an AP of 99.8% for cancer detection on an external dataset.
TransResU-Net: Transformer based ResU-Net for Real-Time Colonoscopy Polyp Segmentation
Jun 17, 2022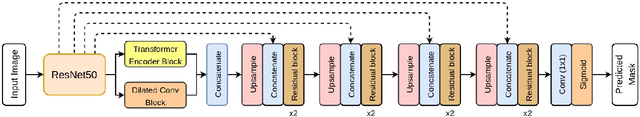
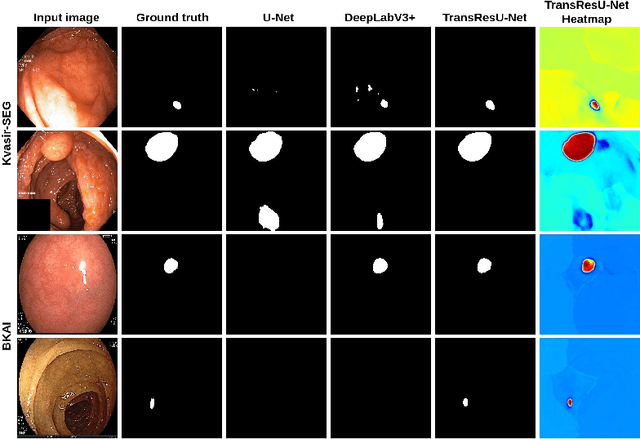
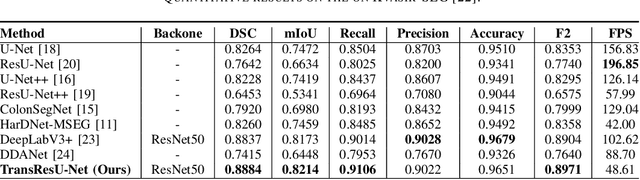
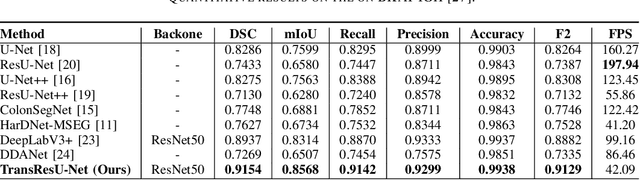
Colorectal cancer (CRC) is one of the most common causes of cancer and cancer-related mortality worldwide. Performing colon cancer screening in a timely fashion is the key to early detection. Colonoscopy is the primary modality used to diagnose colon cancer. However, the miss rate of polyps, adenomas and advanced adenomas remains significantly high. Early detection of polyps at the precancerous stage can help reduce the mortality rate and the economic burden associated with colorectal cancer. Deep learning-based computer-aided diagnosis (CADx) system may help gastroenterologists to identify polyps that may otherwise be missed, thereby improving the polyp detection rate. Additionally, CADx system could prove to be a cost-effective system that improves long-term colorectal cancer prevention. In this study, we proposed a deep learning-based architecture for automatic polyp segmentation, called Transformer ResU-Net (TransResU-Net). Our proposed architecture is built upon residual blocks with ResNet-50 as the backbone and takes the advantage of transformer self-attention mechanism as well as dilated convolution(s). Our experimental results on two publicly available polyp segmentation benchmark datasets showed that TransResU-Net obtained a highly promising dice score and a real-time speed. With high efficacy in our performance metrics, we concluded that TransResU-Net could be a strong benchmark for building a real-time polyp detection system for the early diagnosis, treatment, and prevention of colorectal cancer. The source code of the proposed TransResU-Net is publicly available at https://github.com/nikhilroxtomar/TransResUNet.
Machine Learning Applications in Diagnosis, Treatment and Prognosis of Lung Cancer
Mar 05, 2022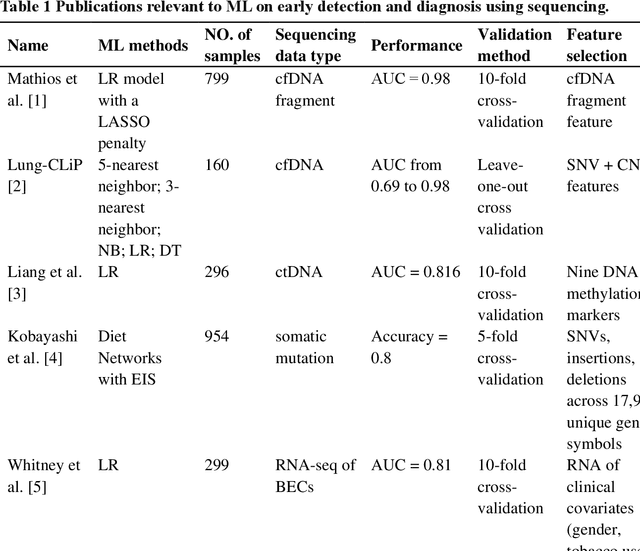
The recent development of imaging and sequencing technologies enables systematic advances in the clinical study of lung cancer. Meanwhile, the human mind is limited in effectively handling and fully utilizing the accumulation of such enormous amounts of data. Machine learning-based approaches play a critical role in integrating and analyzing these large and complex datasets, which have extensively characterized lung cancer through the use of different perspectives from these accrued data. In this article, we provide an overview of machine learning-based approaches that strengthen the varying aspects of lung cancer diagnosis and therapy, including early detection, auxiliary diagnosis, prognosis prediction and immunotherapy practice. Moreover, we highlight the challenges and opportunities for future applications of machine learning in lung cancer.
Detection and Classification of Breast Cancer Metastates Based on U-Net
Sep 09, 2019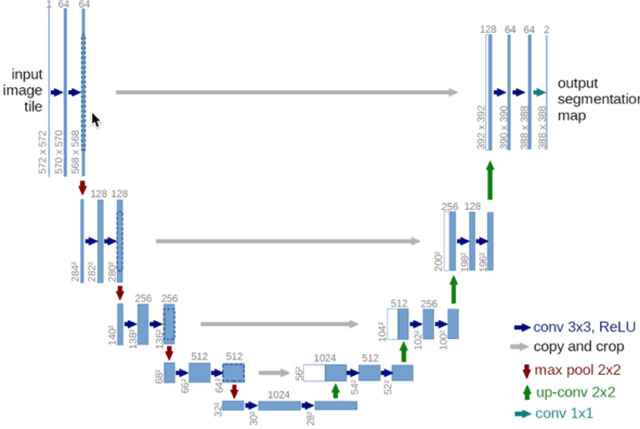
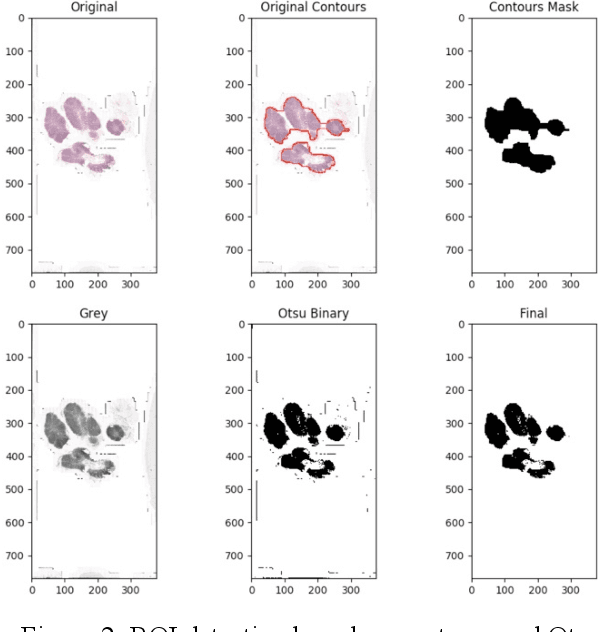
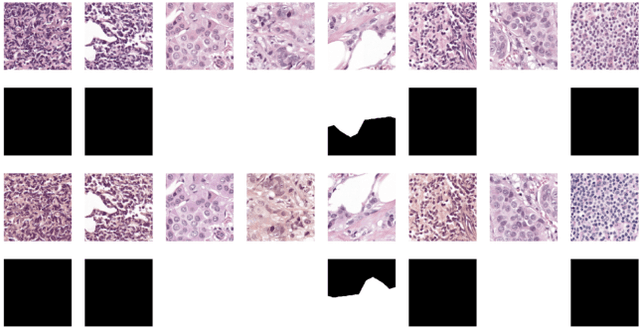
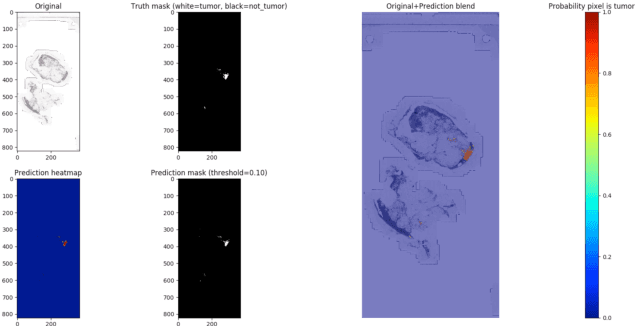
This paper presents U-net based breast cancer metastases detection and classification in lymph nodes, as well as patient-level classification based on metastases detection. The whole pipeline can be divided into five steps: preprocessing and data argumentation, patch-based segmentation, post processing, slide-level classification, and patient-level classification. In order to reduce overfitting and speedup convergence, we applied batch normalization and dropout into U-Net. The final Kappa score reaches 0.902 on training data.
Photonic single perceptron at Giga-OP/s speeds with Kerr microcombs for scalable optical neural networks
May 12, 2021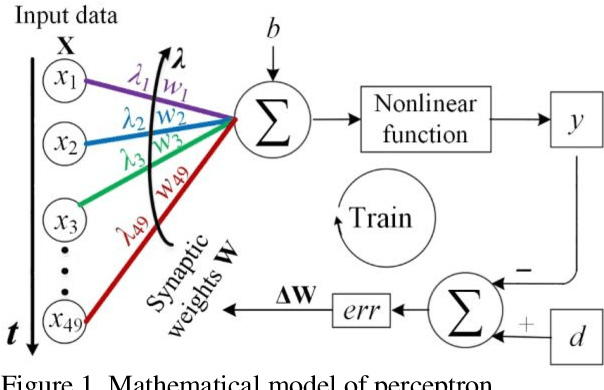
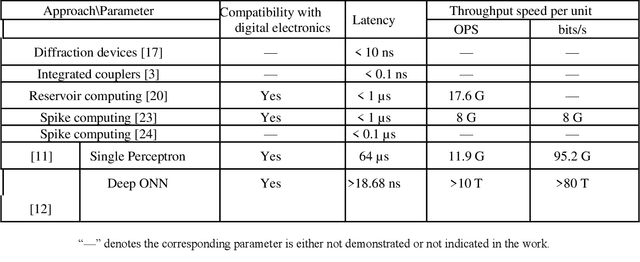
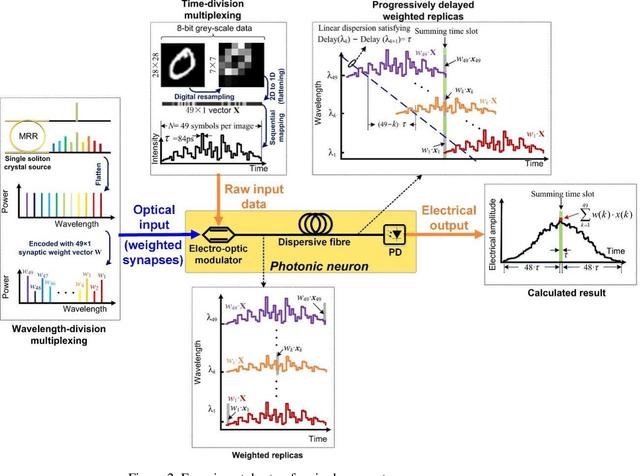
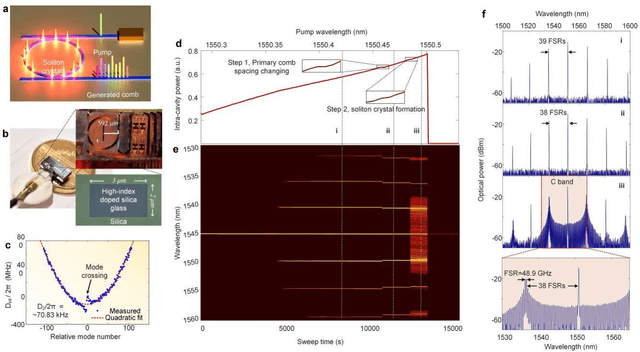
Optical artificial neural networks (ONNs) have significant potential for ultra-high computing speed and energy efficiency. We report a novel approach to ONNs that uses integrated Kerr optical microcombs. This approach is programmable and scalable and is capable of reaching ultrahigh speeds. We demonstrate the basic building block ONNs, a single neuron perceptron, by mapping synapses onto 49 wavelengths to achieve an operating speed of 11.9 x 109 operations per second, or GigaOPS, at 8 bits per operation, which equates to 95.2 gigabits/s (Gbps). We test the perceptron on handwritten digit recognition and cancer cell detection, achieving over 90% and 85% accuracy, respectively. By scaling the perceptron to a deep learning network using off the shelf telecom technology we can achieve high throughput operation for matrix multiplication for real-time massive data processing.
* 14 pages, 8 figures, 107 references. arXiv admin note: substantial text overlap with arXiv:2101.12356
Machine Intelligence-Driven Classification of Cancer Patients-Derived Extracellular Vesicles using Fluorescence Correlation Spectroscopy: Results from a Pilot Study
Feb 01, 2022Patient-derived extracellular vesicles (EVs) that contains a complex biological cargo is a valuable source of liquid biopsy diagnostics to aid in early detection, cancer screening, and precision nanotherapeutics. In this study, we predicted that coupling cancer patient blood-derived EVs to time-resolved spectroscopy and artificial intelligence (AI) could provide a robust cancer screening and follow-up tools. Methods: Fluorescence correlation spectroscopy (FCS) measurements were performed on 24 blood samples-derived EVs. Blood samples were obtained from 15 cancer patients (presenting 5 different types of cancers), and 9 healthy controls (including patients with benign lesions). The obtained FCS autocorrelation spectra were processed into power spectra using the Fast-Fourier Transform algorithm and subjected to various machine learning algorithms to distinguish cancer spectra from healthy control spectra. Results and Applications: The performance of AdaBoost Random Forest (RF) classifier, support vector machine, and multilayer perceptron, were tested on selected frequencies in the N=118 power spectra. The RF classifier exhibited a 90% classification accuracy and high sensitivity and specificity in distinguishing the FCS power spectra of cancer patients from those of healthy controls. Further, an image convolutional neural network (CNN), ResNet network, and a quantum CNN were assessed on the power spectral images as additional validation tools. All image-based CNNs exhibited a nearly equal classification performance with an accuracy of roughly 82% and reasonably high sensitivity and specificity scores. Our pilot study demonstrates that AI-algorithms coupled to time-resolved FCS power spectra can accurately and differentially classify the complex patient-derived EVs from different cancer samples of distinct tissue subtypes.
Multi-level colonoscopy malignant tissue detection with adversarial CAC-UNet
Jun 30, 2020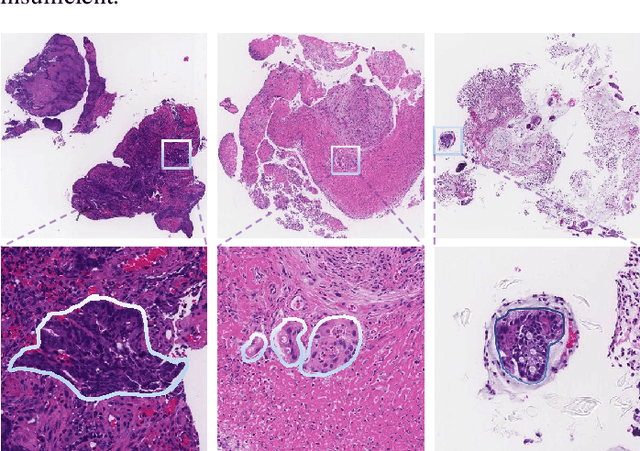

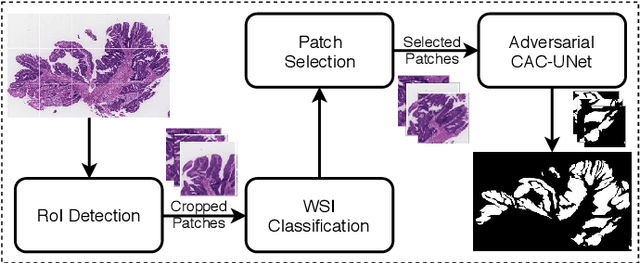
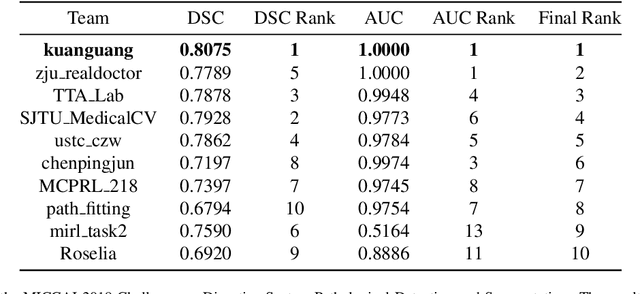
The automatic and objective medical diagnostic model can be valuable to achieve early cancer detection, and thus reducing the mortality rate. In this paper, we propose a highly efficient multi-level malignant tissue detection through the designed adversarial CAC-UNet. A patch-level model with a pre-prediction strategy and a malignancy area guided label smoothing is adopted to remove the negative WSIs, with which to lower the risk of false positive detection. For the selected key patches by multi-model ensemble, an adversarial context-aware and appearance consistency UNet (CAC-UNet) is designed to achieve robust segmentation. In CAC-UNet, mirror designed discriminators are able to seamlessly fuse the whole feature maps of the skillfully designed powerful backbone network without any information loss. Besides, a mask prior is further added to guide the accurate segmentation mask prediction through an extra mask-domain discriminator. The proposed scheme achieves the best results in MICCAI DigestPath2019 challenge on colonoscopy tissue segmentation and classification task. The full implementation details and the trained models are available at https://github.com/Raykoooo/CAC-UNet.
End-to-end Prostate Cancer Detection in bpMRI via 3D CNNs: Effect of Attention Mechanisms, Clinical Priori and Decoupled False Positive Reduction
Jan 28, 2021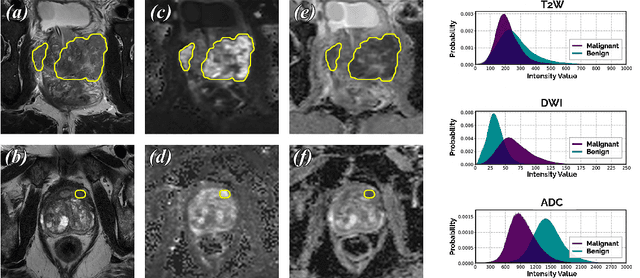
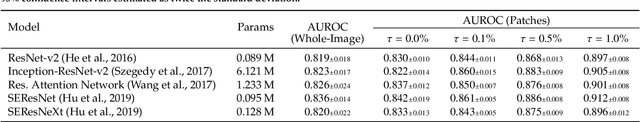
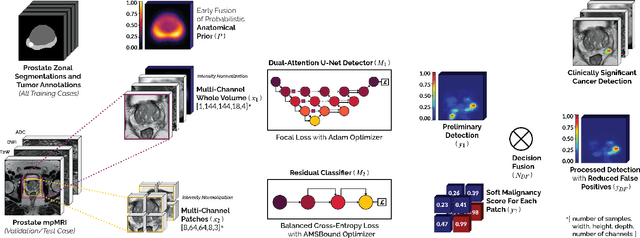
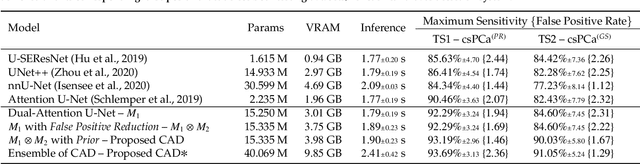
We present a novel multi-stage 3D computer-aided detection and diagnosis (CAD) model for automated localization of clinically significant prostate cancer (csPCa) in bi-parametric MR imaging (bpMRI). Deep attention mechanisms drive its detection network, targeting multi-resolution, salient structures and highly discriminative feature dimensions, in order to accurately identify csPCa lesions from indolent cancer and the wide range of benign pathology that can afflict the prostate gland. In parallel, a decoupled residual classifier is used to achieve consistent false positive reduction, without sacrificing high sensitivity or computational efficiency. Furthermore, a probabilistic anatomical prior, which captures the spatial prevalence of csPCa as well as its zonal distinction, is computed and encoded into the CNN architecture to guide model generalization with domain-specific clinical knowledge. For 486 institutional testing scans, the 3D CAD system achieves $83.69\pm5.22\%$ and $93.19\pm2.96\%$ detection sensitivity at 0.50 and 1.46 false positive(s) per patient, respectively, along with $0.882$ AUROC in patient-based diagnosis $-$significantly outperforming four state-of-the-art baseline architectures (U-SEResNet, UNet++, nnU-Net, Attention U-Net) from recent literature. For 296 external testing scans, the ensembled CAD system shares moderate agreement with a consensus of expert radiologists ($76.69\%$; $kappa=0.511$) and independent pathologists ($81.08\%$; $kappa=0.559$); demonstrating strong generalization to histologically-confirmed malignancies, despite using 1950 training-validation cases with radiologically-estimated annotations only.
Computational Enhancement of Molecularly Targeted Contrast-Enhanced Ultrasound: Application to Human Breast Tumor Imaging
Jun 22, 2020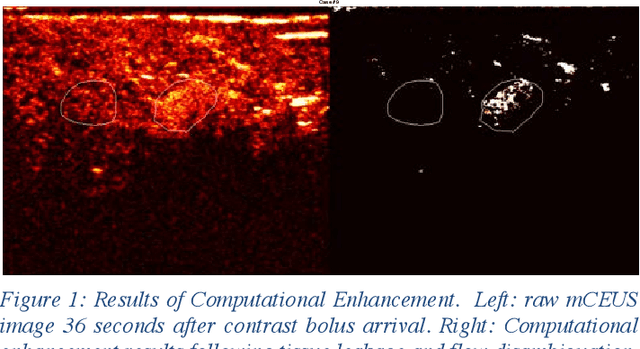
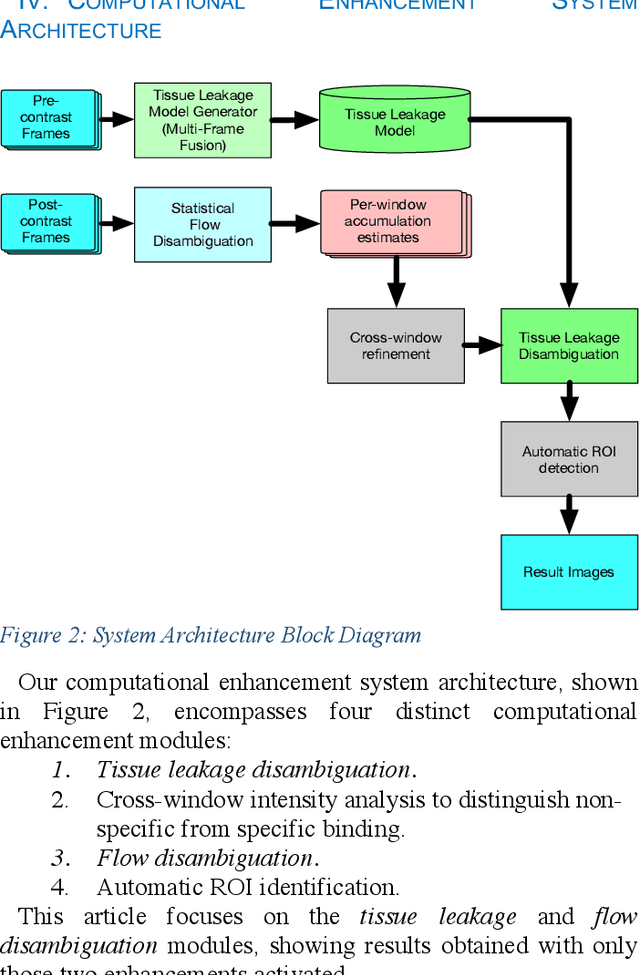
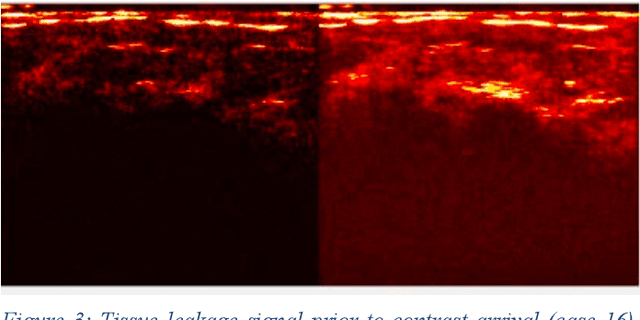
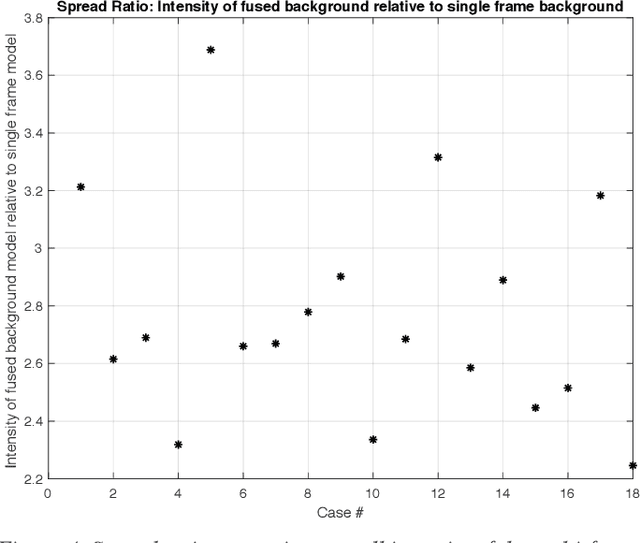
Molecularly targeted contrast enhanced ultrasound (mCEUS) is a clinically promising approach for early cancer detection through targeted imaging of VEGFR2 (KDR) receptors. We have developed computational enhancement techniques for mCEUS tailored to address the unique challenges of imaging contrast accumulation in humans. These techniques utilize dynamic analysis to distinguish molecularly bound contrast agent from other contrast-mode signal sources, enabling analysis of contrast agent accumulation to be performed during contrast bolus arrival when the signal due to molecular binding is strongest. Applied to the 18 human patient examinations of the first-in-human molecular ultrasound breast lesion study, computational enhancement improved the ability to differentiate between pathology-proven lesion and pathology-proven normal tissue in real-world human examination conditions that involved both patient and probe motion, with improvements in contrast ratio between lesion and normal tissue that in most cases exceed an order of magnitude (10x). Notably, computational enhancement eliminated a false positive result in which tissue leakage signal was misinterpreted by radiologists to be contrast agent accumulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge