"cancer detection": models, code, and papers
Mass Segmentation in Automated 3-D Breast Ultrasound Using Dual-Path U-net
Sep 17, 2021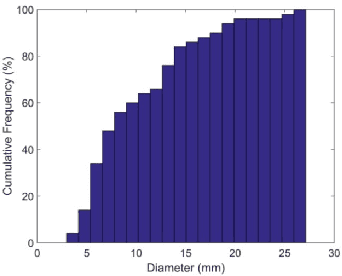
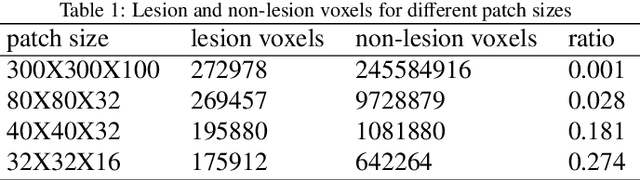
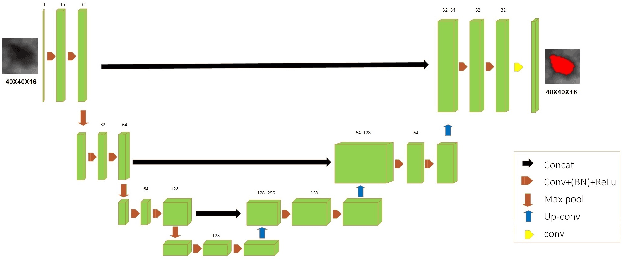
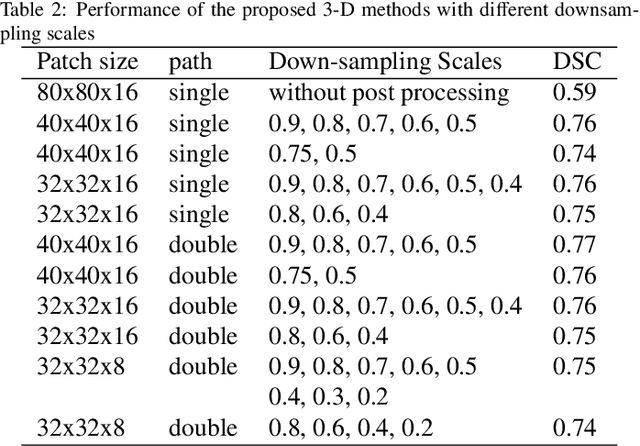
Automated 3-D breast ultrasound (ABUS) is a newfound system for breast screening that has been proposed as a supplementary modality to mammography for breast cancer detection. While ABUS has better performance in dense breasts, reading ABUS images is exhausting and time-consuming. So, a computer-aided detection system is necessary for interpretation of these images. Mass segmentation plays a vital role in the computer-aided detection systems and it affects the overall performance. Mass segmentation is a challenging task because of the large variety in size, shape, and texture of masses. Moreover, an imbalanced dataset makes segmentation harder. A novel mass segmentation approach based on deep learning is introduced in this paper. The deep network that is used in this study for image segmentation is inspired by U-net, which has been used broadly for dense segmentation in recent years. The system's performance was determined using a dataset of 50 masses including 38 malign and 12 benign lesions. The proposed segmentation method attained a mean Dice of 0.82 which outperformed a two-stage supervised edge-based method with a mean Dice of 0.74 and an adaptive region growing method with a mean Dice of 0.65.
Pricing Algorithmic Insurance
Jun 01, 2021
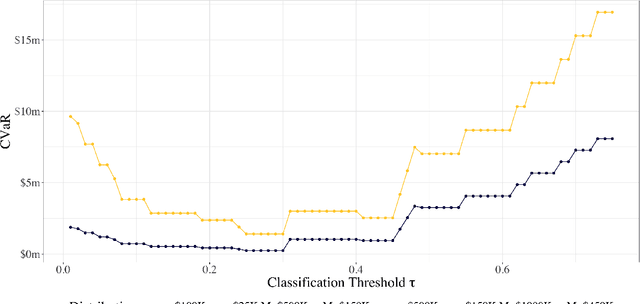
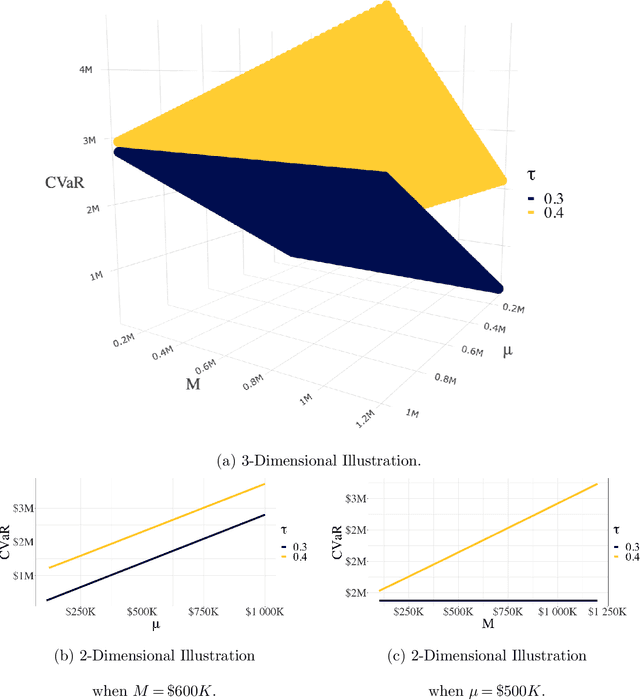
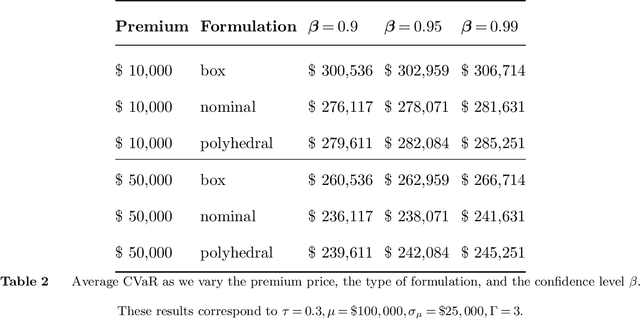
As machine learning algorithms start to get integrated into the decision-making process of companies and organizations, insurance products will be developed to protect their owners from risk. We introduce the concept of algorithmic insurance and present a quantitative framework to enable the pricing of the derived insurance contracts. We propose an optimization formulation to estimate the risk exposure and price for a binary classification model. Our approach outlines how properties of the model, such as accuracy, interpretability and generalizability, can influence the insurance contract evaluation. To showcase a practical implementation of the proposed framework, we present a case study of medical malpractice in the context of breast cancer detection. Our analysis focuses on measuring the effect of the model parameters on the expected financial loss and identifying the aspects of algorithmic performance that predominantly affect the price of the contract.
Bridging the gap between prostate radiology and pathology through machine learning
Dec 03, 2021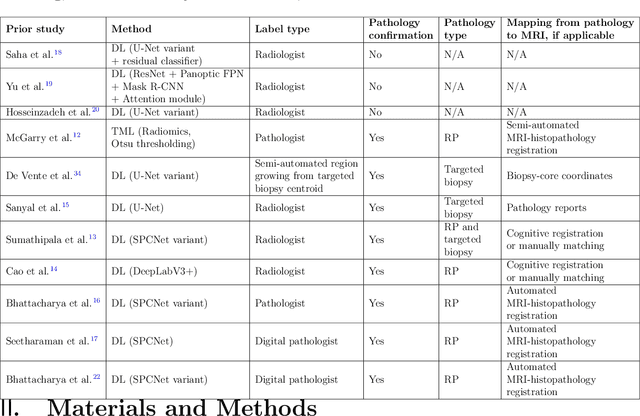
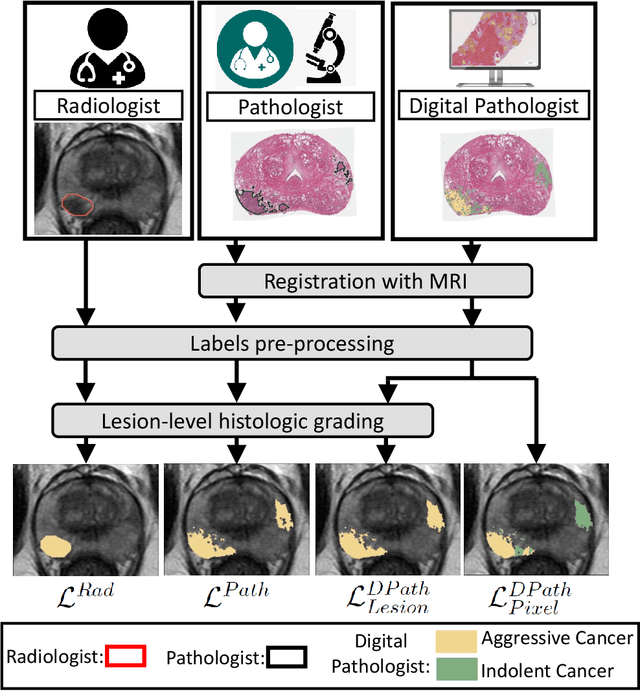
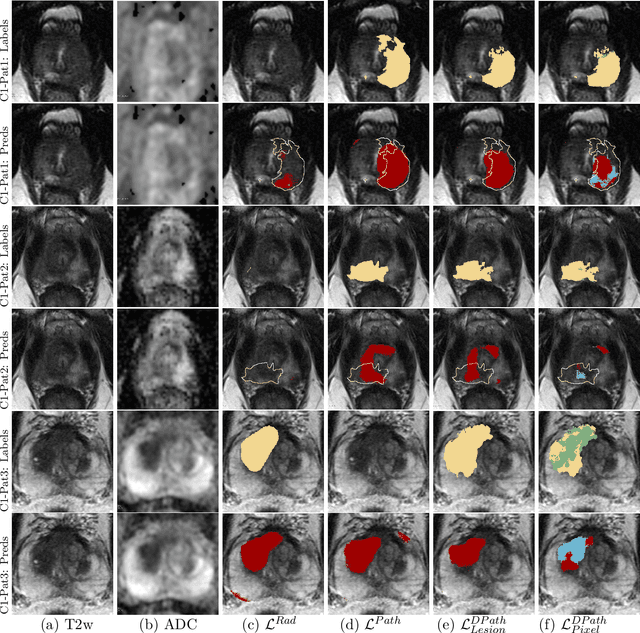
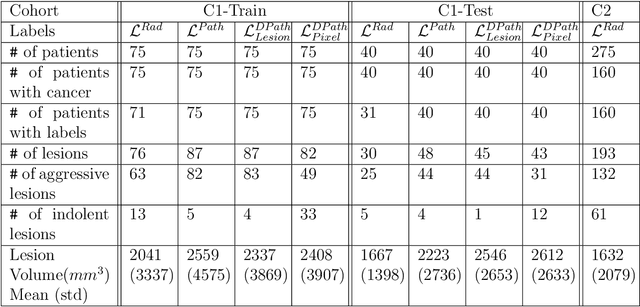
Prostate cancer is the second deadliest cancer for American men. While Magnetic Resonance Imaging (MRI) is increasingly used to guide targeted biopsies for prostate cancer diagnosis, its utility remains limited due to high rates of false positives and false negatives as well as low inter-reader agreements. Machine learning methods to detect and localize cancer on prostate MRI can help standardize radiologist interpretations. However, existing machine learning methods vary not only in model architecture, but also in the ground truth labeling strategies used for model training. In this study, we compare different labeling strategies, namely, pathology-confirmed radiologist labels, pathologist labels on whole-mount histopathology images, and lesion-level and pixel-level digital pathologist labels (previously validated deep learning algorithm on histopathology images to predict pixel-level Gleason patterns) on whole-mount histopathology images. We analyse the effects these labels have on the performance of the trained machine learning models. Our experiments show that (1) radiologist labels and models trained with them can miss cancers, or underestimate cancer extent, (2) digital pathologist labels and models trained with them have high concordance with pathologist labels, and (3) models trained with digital pathologist labels achieve the best performance in prostate cancer detection in two different cohorts with different disease distributions, irrespective of the model architecture used. Digital pathologist labels can reduce challenges associated with human annotations, including labor, time, inter- and intra-reader variability, and can help bridge the gap between prostate radiology and pathology by enabling the training of reliable machine learning models to detect and localize prostate cancer on MRI.
Encoding Clinical Priori in 3D Convolutional Neural Networks for Prostate Cancer Detection in bpMRI
Nov 03, 2020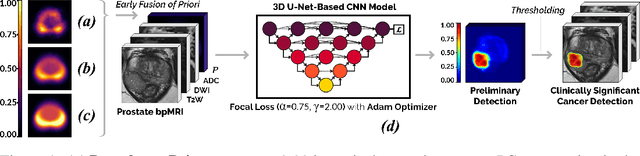
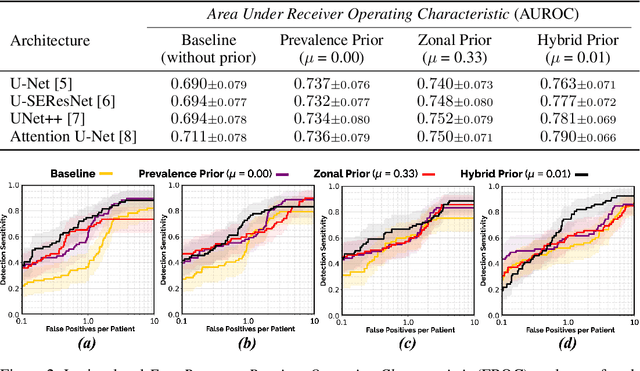
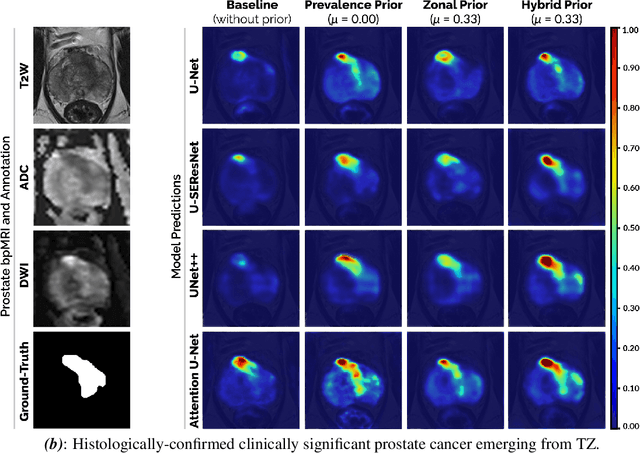
We hypothesize that anatomical priors can be viable mediums to infuse domain-specific clinical knowledge into state-of-the-art convolutional neural networks (CNN) based on the U-Net architecture. We introduce a probabilistic population prior which captures the spatial prevalence and zonal distinction of clinically significant prostate cancer (csPCa), in order to improve its computer-aided detection (CAD) in bi-parametric MR imaging (bpMRI). To evaluate performance, we train 3D adaptations of the U-Net, U-SEResNet, UNet++ and Attention U-Net using 800 institutional training-validation scans, paired with radiologically-estimated annotations and our computed prior. For 200 independent testing bpMRI scans with histologically-confirmed delineations of csPCa, our proposed method of encoding clinical priori demonstrates a strong ability to improve patient-based diagnosis (upto 8.70% increase in AUROC) and lesion-level detection (average increase of 1.08 pAUC between 0.1-1.0 false positive per patient) across all four architectures.
Capsule GAN for Prostate MRI Super-Resolution
May 20, 2021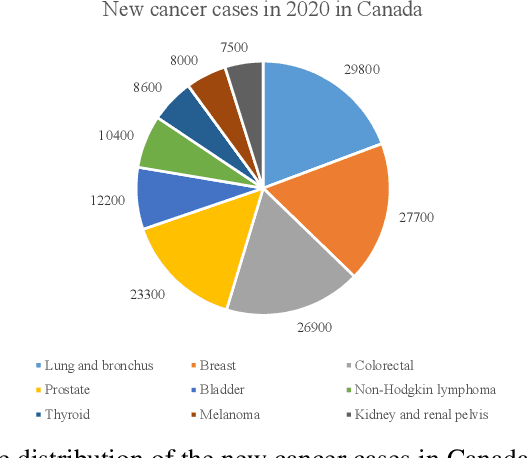
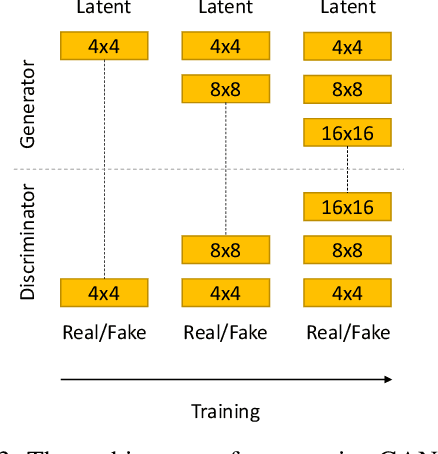
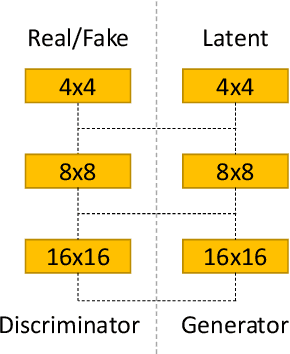
Prostate cancer is a very common disease among adult men. One in seven Canadian men is diagnosed with this cancer in their lifetime. Super-Resolution (SR) can facilitate early diagnosis and potentially save many lives. In this paper, a robust and accurate model is proposed for prostate MRI SR. The model is trained on the Prostate-Diagnosis and PROSTATEx datasets. The proposed model outperformed the state-of-the-art prostate SR model in all similarity metrics with notable margins. A new task-specific similarity assessment is introduced as well. A classifier is trained for severe cancer detection and the drop in the accuracy of this model when dealing with super-resolved images is used for evaluating the ability of medical detail reconstruction of the SR models. The proposed SR model is a step towards an efficient and accurate general medical SR platform.
The Fast and Accurate Approach to Detection and Segmentation of Melanoma Skin Cancer using Fine-tuned Yolov3 and SegNet Based on Deep Transfer Learning
Oct 11, 2022
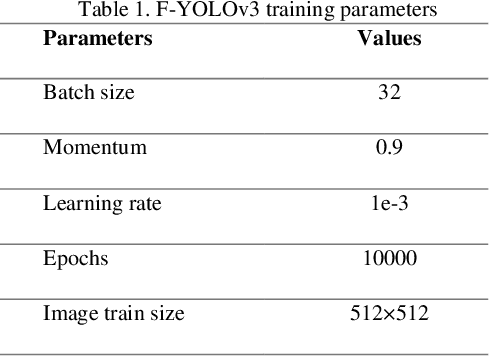
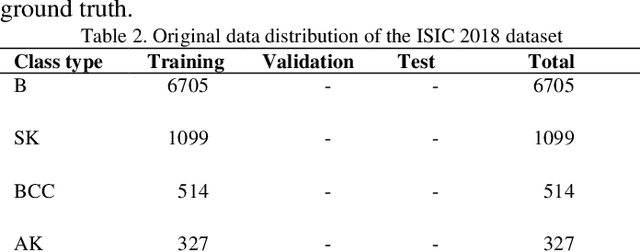
Melanoma is one of the most serious skin cancers that can occur in any part of the human skin. Early diagnosing melanoma lesions will significantly increase their chances of being cured. Improving melanoma segmentation will help doctors or surgical robots remove the lesion more accurately from body parts. Recently, the learning-based segmentation methods achieved desired results in image segmentation compared to traditional algorithms. This study proposes a new method to improve melanoma skin lesions detection and segmentation by defining a two-step pipeline based on deep learning models. Our methods were evaluated on ISIC 2018 (Skin Lesion Analysis Towards Melanoma Detection Challenge Dataset) well-known dataset. The proposed methods consist of two main parts for real-time detection of lesion location and segmentation. In the detection section, the location of the skin lesion is precisely detected by the fine-tuned You Only Look Once version 3 (F-YOLOv3) and then fed into the fine-tuned Segmentation Network (F-SegNet). Skin lesion localization helps to reduce the unnecessary calculation of whole images for segmentation. The results show that our proposed F-YOLOv3 achieves better performance as 96% in mAP. Compared to state-of-the-art segmentation approaches, our F-SegNet achieves higher performance for accuracy, dice coefficient, and Jaccard index at 95.16%, 92.81%, and 86.2%, respectively.
Out of distribution detection for skin and malaria images
Nov 02, 2021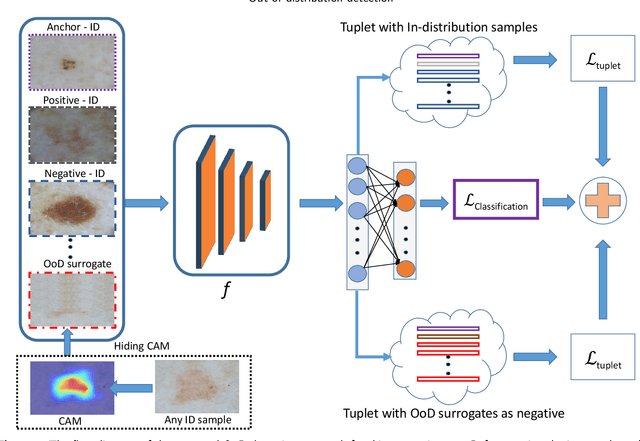
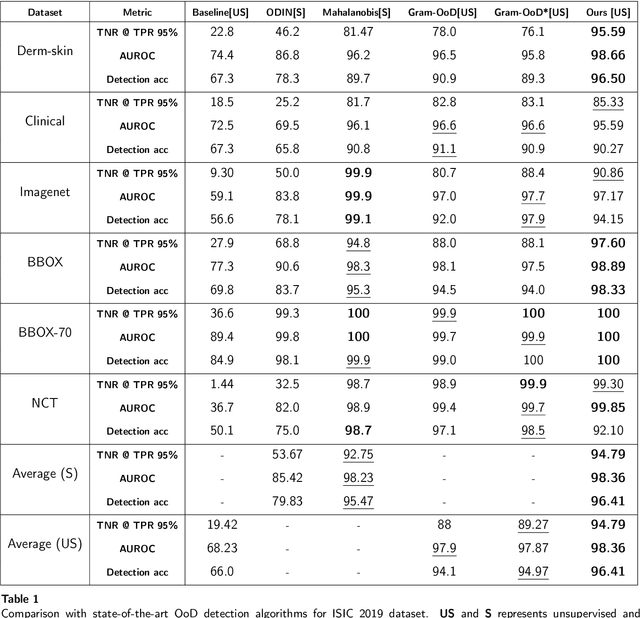
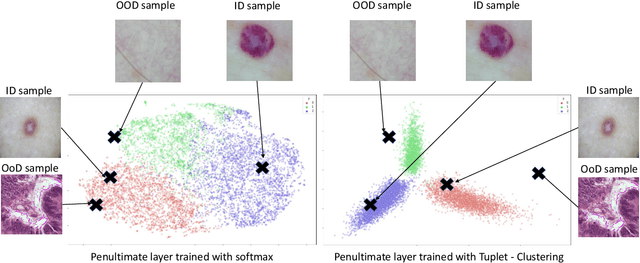
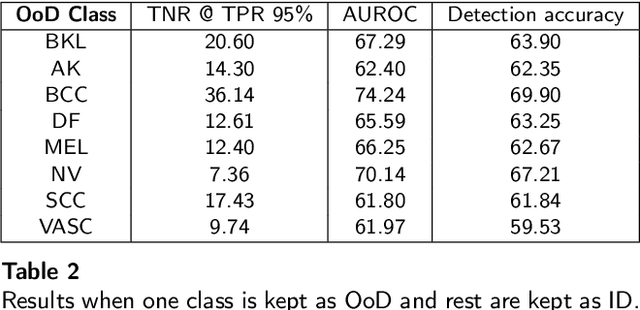
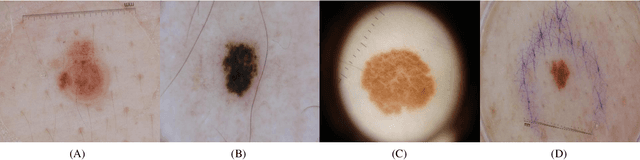
Deep neural networks have shown promising results in disease detection and classification using medical image data. However, they still suffer from the challenges of handling real-world scenarios especially reliably detecting out-of-distribution (OoD) samples. We propose an approach to robustly classify OoD samples in skin and malaria images without the need to access labeled OoD samples during training. Specifically, we use metric learning along with logistic regression to force the deep networks to learn much rich class representative features. To guide the learning process against the OoD examples, we generate ID similar-looking examples by either removing class-specific salient regions in the image or permuting image parts and distancing them away from in-distribution samples. During inference time, the K-reciprocal nearest neighbor is employed to detect out-of-distribution samples. For skin cancer OoD detection, we employ two standard benchmark skin cancer ISIC datasets as ID, and six different datasets with varying difficulty levels were taken as out of distribution. For malaria OoD detection, we use the BBBC041 malaria dataset as ID and five different challenging datasets as out of distribution. We achieved state-of-the-art results, improving 5% and 4% in TNR@TPR95% over the previous state-of-the-art for skin cancer and malaria OoD detection respectively.
Breast Cancer Detection Using Convolutional Neural Networks
Mar 19, 2020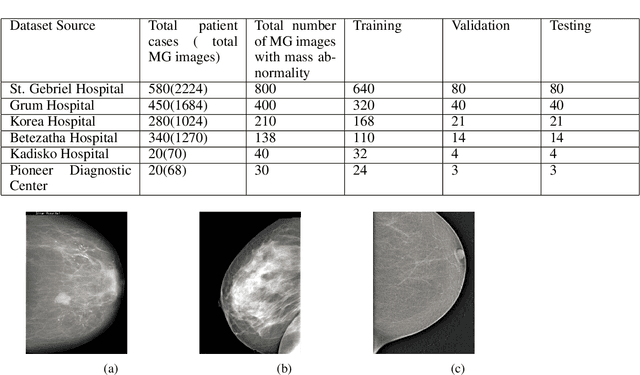
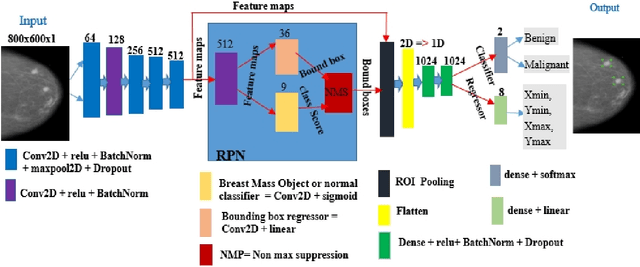

Breast cancer is prevalent in Ethiopia that accounts 34% among women cancer patients. The diagnosis technique in Ethiopia is manual which was proven to be tedious, subjective, and challenging. Deep learning techniques are revolutionizing the field of medical image analysis and hence in this study, we proposed Convolutional Neural Networks (CNNs) for breast mass detection so as to minimize the overheads of manual analysis. CNN architecture is designed for the feature extraction stage and adapted both the Region Proposal Network (RPN) and Region of Interest (ROI) portion of the faster R-CNN for the automated breast mass abnormality detection. Our model detects mass region and classifies them into benign or malignant abnormality in mammogram(MG) images at once. For the proposed model, MG images were collected from different hospitals, locally.The images were passed through different preprocessing stages such as gaussian filter, median filter, bilateral filters and extracted the region of the breast from the background of the MG image. The performance of the model on test dataset is found to be: detection accuracy 91.86%, sensitivity of 94.67% and AUC-ROC of 92.2%.
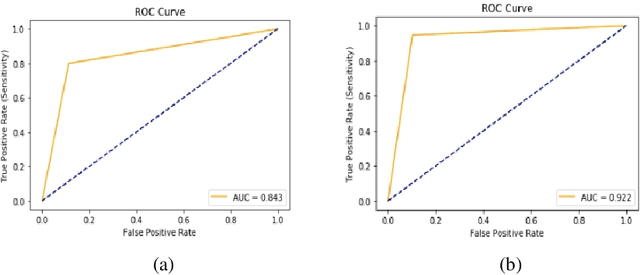
Surpassing the Human Accuracy: Detecting Gallbladder Cancer from USG Images with Curriculum Learning
Apr 25, 2022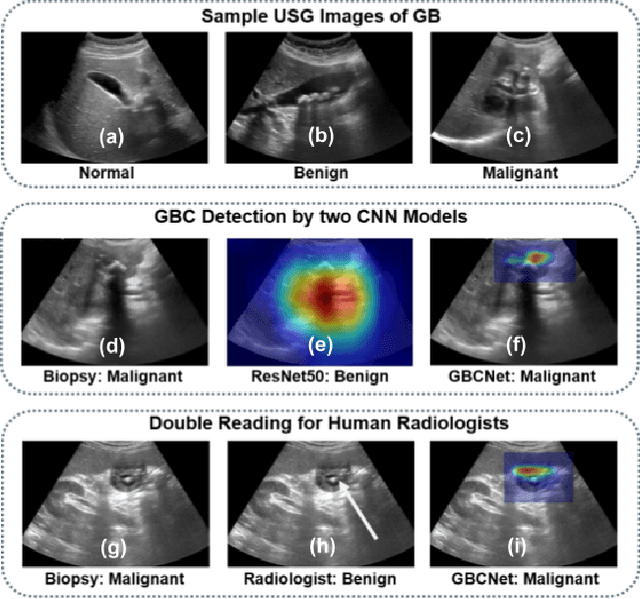
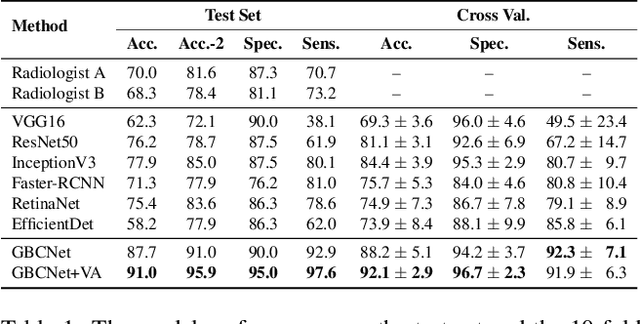
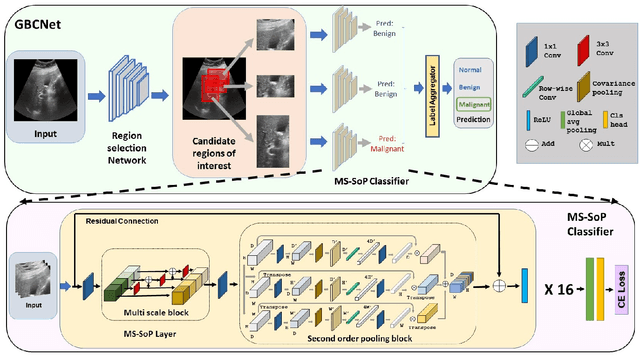
We explore the potential of CNN-based models for gallbladder cancer (GBC) detection from ultrasound (USG) images as no prior study is known. USG is the most common diagnostic modality for GB diseases due to its low cost and accessibility. However, USG images are challenging to analyze due to low image quality, noise, and varying viewpoints due to the handheld nature of the sensor. Our exhaustive study of state-of-the-art (SOTA) image classification techniques for the problem reveals that they often fail to learn the salient GB region due to the presence of shadows in the USG images. SOTA object detection techniques also achieve low accuracy because of spurious textures due to noise or adjacent organs. We propose GBCNet to tackle the challenges in our problem. GBCNet first extracts the regions of interest (ROIs) by detecting the GB (and not the cancer), and then uses a new multi-scale, second-order pooling architecture specializing in classifying GBC. To effectively handle spurious textures, we propose a curriculum inspired by human visual acuity, which reduces the texture biases in GBCNet. Experimental results demonstrate that GBCNet significantly outperforms SOTA CNN models, as well as the expert radiologists. Our technical innovations are generic to other USG image analysis tasks as well. Hence, as a validation, we also show the efficacy of GBCNet in detecting breast cancer from USG images. Project page with source code, trained models, and data is available at https://gbc-iitd.github.io/gbcnet
Demystifying the Transferability of Adversarial Attacks in Computer Networks
Oct 09, 2021
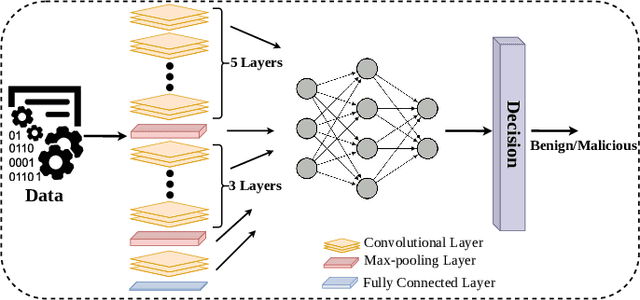
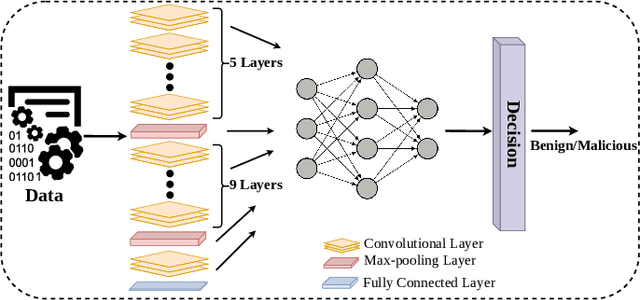
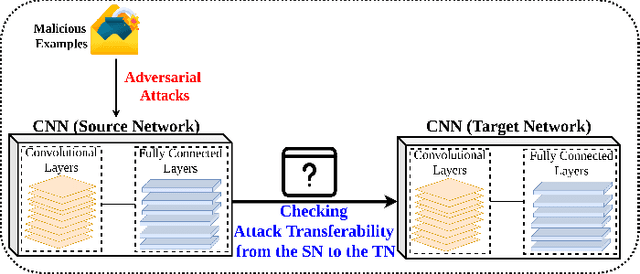
Deep Convolutional Neural Networks (CNN) models are one of the most popular networks in deep learning. With their large fields of application in different areas, they are extensively used in both academia and industry. CNN-based models include several exciting implementations such as early breast cancer detection or detecting developmental delays in children (e.g., autism, speech disorders, etc.). However, previous studies demonstrate that these models are subject to various adversarial attacks. Interestingly, some adversarial examples could potentially still be effective against different unknown models. This particular property is known as adversarial transferability, and prior works slightly analyzed this characteristic in a very limited application domain. In this paper, we aim to demystify the transferability threats in computer networks by studying the possibility of transferring adversarial examples. In particular, we provide the first comprehensive study which assesses the robustness of CNN-based models for computer networks against adversarial transferability. In our experiments, we consider five different attacks: (1) the Iterative Fast Gradient Method (I-FGSM), (2) the Jacobian-based Saliency Map attack (JSMA), (3) the L-BFGS attack, (4) the Projected Gradient Descent attack (PGD), and (5) the DeepFool attack. These attacks are performed against two well-known datasets: the N-BaIoT dataset and the Domain Generating Algorithms (DGA) dataset. Our results show that the transferability happens in specific use cases where the adversary can easily compromise the victim's network with very few knowledge of the targeted model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge