"cancer detection": models, code, and papers
Recent trends and analysis of Generative Adversarial Networks in Cervical Cancer Imaging
Sep 23, 2022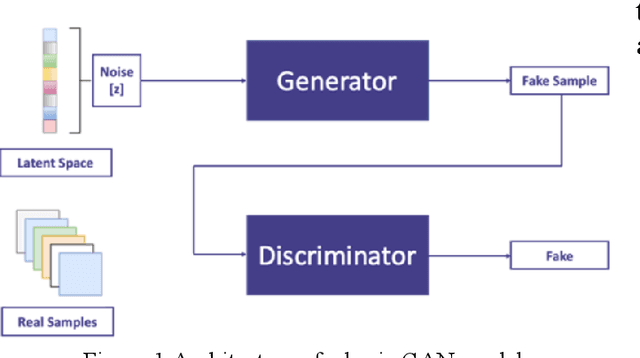
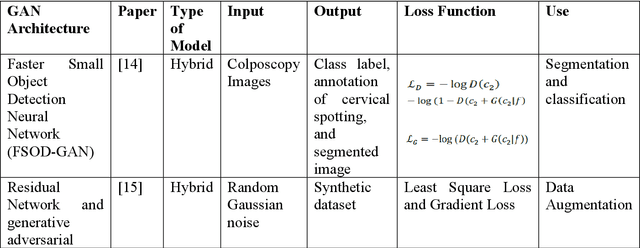
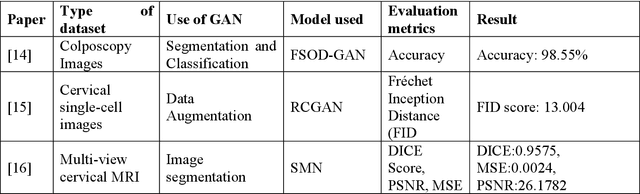
Cervical cancer is one of the most common types of cancer found in females. It contributes to 6-29% of all cancers in women. It is caused by the Human Papilloma Virus (HPV). The 5-year survival chances of cervical cancer range from 17%-92% depending upon the stage at which it is detected. Early detection of this disease helps in better treatment and survival rate of the patient. Many deep learning algorithms are being used for the detection of cervical cancer these days. A special category of deep learning techniques known as Generative Adversarial Networks (GANs) are catching up with speed in the screening, detection, and classification of cervical cancer. In this work, we present a detailed analysis of the recent trends relating to the use of various GAN models, their applications, and the evaluation metrics used for their performance evaluation in the field of cervical cancer imaging.
* 7 pages, proceedings of ICCS 2022 (https://www.cecmohali.org/iccs2022/), published in neuroquantology (https://www.neuroquantology.com/article.php?id=7222 , DOI:10.14704/nq.2022.20.9.NQ44381 )
A Review of Generative Adversarial Networks in Cancer Imaging: New Applications, New Solutions
Jul 20, 2021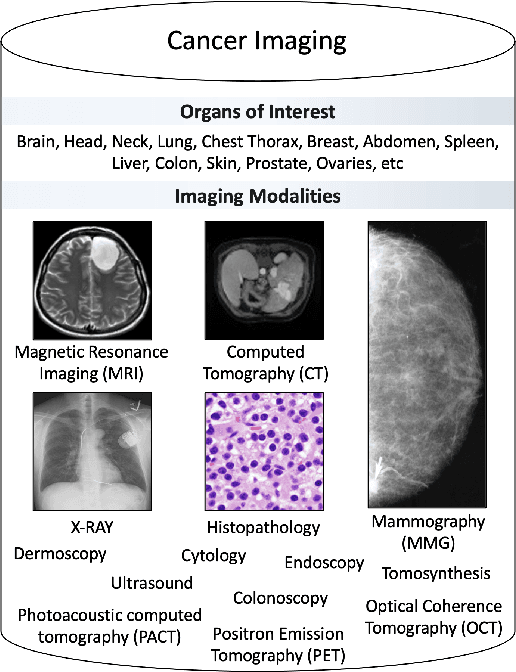
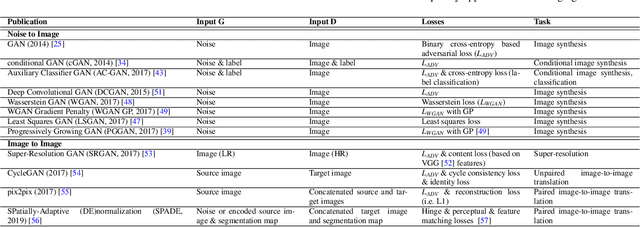
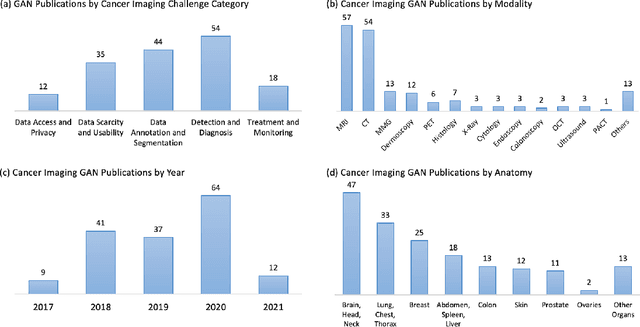
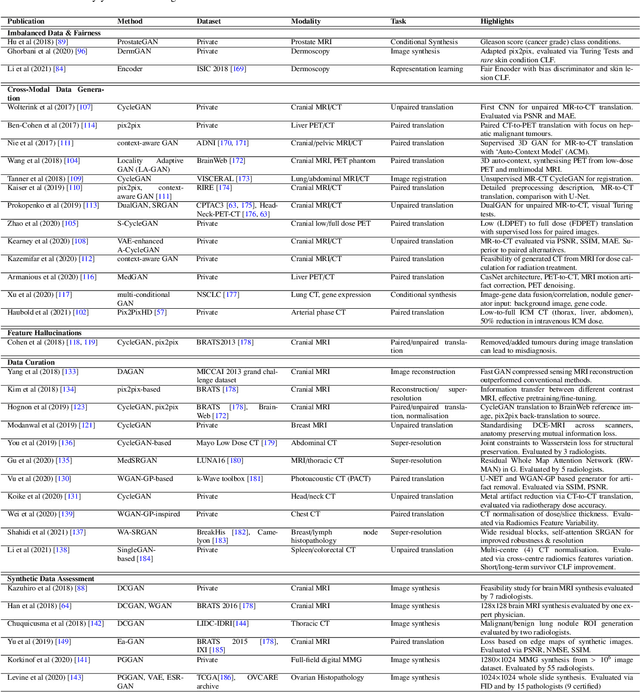
Despite technological and medical advances, the detection, interpretation, and treatment of cancer based on imaging data continue to pose significant challenges. These include high inter-observer variability, difficulty of small-sized lesion detection, nodule interpretation and malignancy determination, inter- and intra-tumour heterogeneity, class imbalance, segmentation inaccuracies, and treatment effect uncertainty. The recent advancements in Generative Adversarial Networks (GANs) in computer vision as well as in medical imaging may provide a basis for enhanced capabilities in cancer detection and analysis. In this review, we assess the potential of GANs to address a number of key challenges of cancer imaging, including data scarcity and imbalance, domain and dataset shifts, data access and privacy, data annotation and quantification, as well as cancer detection, tumour profiling and treatment planning. We provide a critical appraisal of the existing literature of GANs applied to cancer imagery, together with suggestions on future research directions to address these challenges. We analyse and discuss 163 papers that apply adversarial training techniques in the context of cancer imaging and elaborate their methodologies, advantages and limitations. With this work, we strive to bridge the gap between the needs of the clinical cancer imaging community and the current and prospective research on GANs in the artificial intelligence community.
Transfer learning for cancer diagnosis in histopathological images
Dec 31, 2021
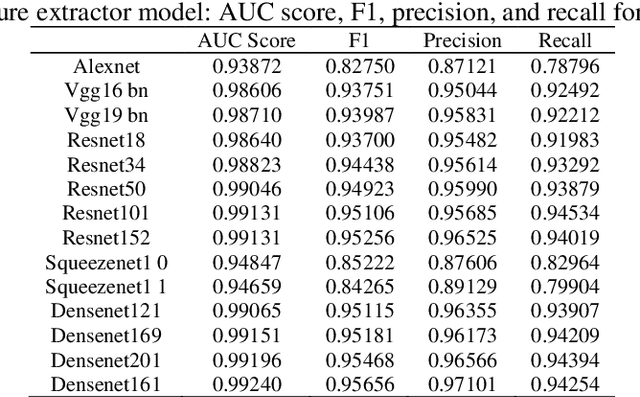
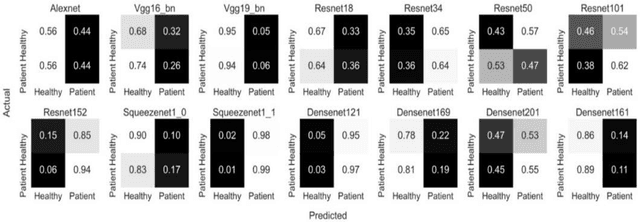
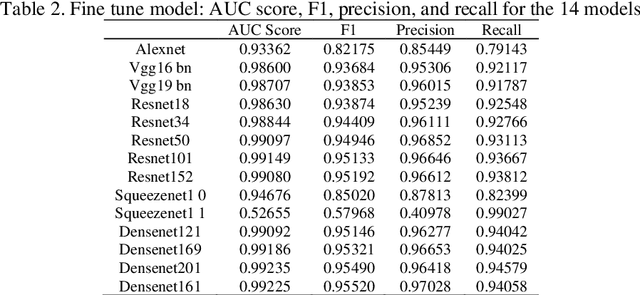
Transfer learning allows us to exploit knowledge gained from one task to assist in solving another but relevant task. In modern computer vision research, the question is which architecture performs better for a given dataset. In this paper, we compare the performance of 14 pre-trained ImageNet models on the histopathologic cancer detection dataset, where each model has been configured as a naive model, feature extractor model, or fine-tuned model. Densenet161 has been shown to have high precision whilst Resnet101 has a high recall. A high precision model is suitable to be used when follow-up examination cost is high, whilst low precision but a high recall/sensitivity model can be used when the cost of follow-up examination is low. Results also show that transfer learning helps to converge a model faster.
Detecting Bone Lesions in X-Ray Under Diverse Acquisition Conditions
Dec 15, 2022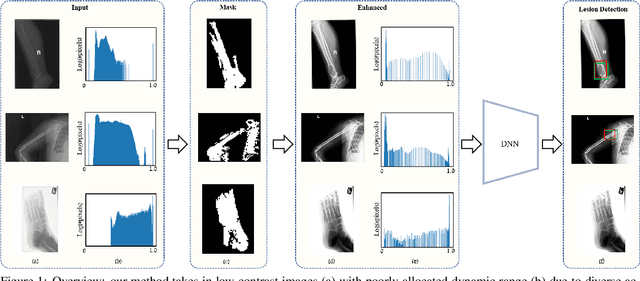
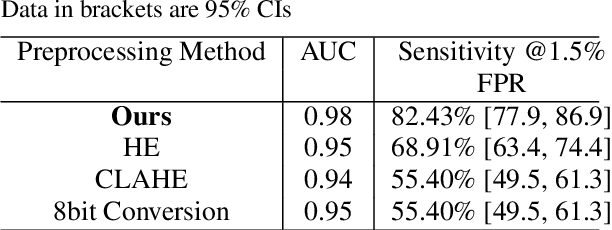
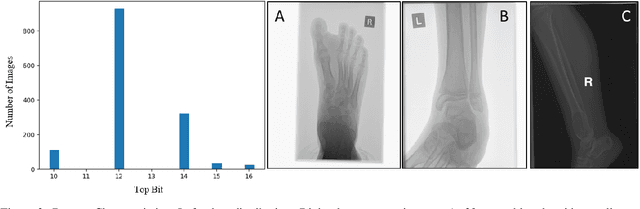
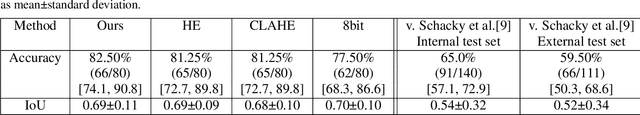
The diagnosis of primary bone tumors is challenging, as the initial complaints are often non-specific. Early detection of bone cancer is crucial for a favorable prognosis. Incidentally, lesions may be found on radiographs obtained for other reasons. However, these early indications are often missed. In this work, we propose an automatic algorithm to detect bone lesions in conventional radiographs to facilitate early diagnosis. Detecting lesions in such radiographs is challenging: first, the prevalence of bone cancer is very low; any method must show high precision to avoid a prohibitive number of false alarms. Second, radiographs taken in health maintenance organizations (HMOs) or emergency departments (EDs) suffer from inherent diversity due to different X-ray machines, technicians and imaging protocols. This diversity poses a major challenge to any automatic analysis method. We propose to train an off-the-shelf object detection algorithm to detect lesions in radiographs. The novelty of our approach stems from a dedicated preprocessing stage that directly addresses the diversity of the data. The preprocessing consists of self-supervised region-of-interest detection using vision transformer (ViT), and a foreground-based histogram equalization for contrast enhancement to relevant regions only. We evaluate our method via a retrospective study that analyzes bone tumors on radiographs acquired from January 2003 to December 2018 under diverse acquisition protocols. Our method obtains 82.43% sensitivity at 1.5% false-positive rate and surpasses existing preprocessing methods. For lesion detection, our method achieves 82.5% accuracy and an IoU of 0.69. The proposed preprocessing method enables to effectively cope with the inherent diversity of radiographs acquired in HMOs and EDs.
A Comparative Analysis of Transfer Learning-based Techniques for the Classification of Melanocytic Nevi
Nov 20, 2022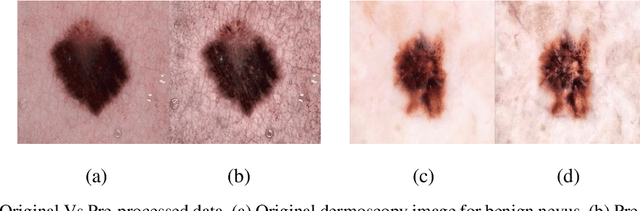

Skin cancer is a fatal manifestation of cancer. Unrepaired deoxyribo-nucleic acid (DNA) in skin cells, causes genetic defects in the skin and leads to skin cancer. To deal with lethal mortality rates coupled with skyrocketing costs of medical treatment, early diagnosis is mandatory. To tackle these challenges, researchers have developed a variety of rapid detection tools for skin cancer. Lesion-specific criteria are utilized to distinguish benign skin cancer from malignant melanoma. In this study, a comparative analysis has been performed on five Transfer Learning-based techniques that have the potential to be leveraged for the classification of melanocytic nevi. These techniques are based on deep convolutional neural networks (DCNNs) that have been pre-trained on thousands of open-source images and are used for day-to-day classification tasks in many instances.
Image Data collection and implementation of deep learning-based model in detecting Monkeypox disease using modified VGG16
Jun 04, 2022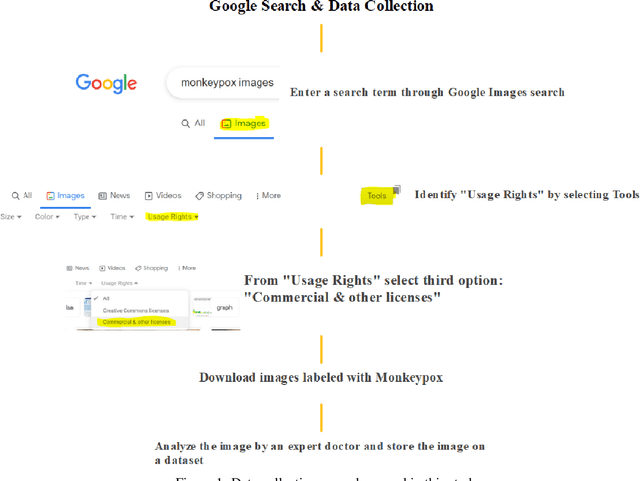
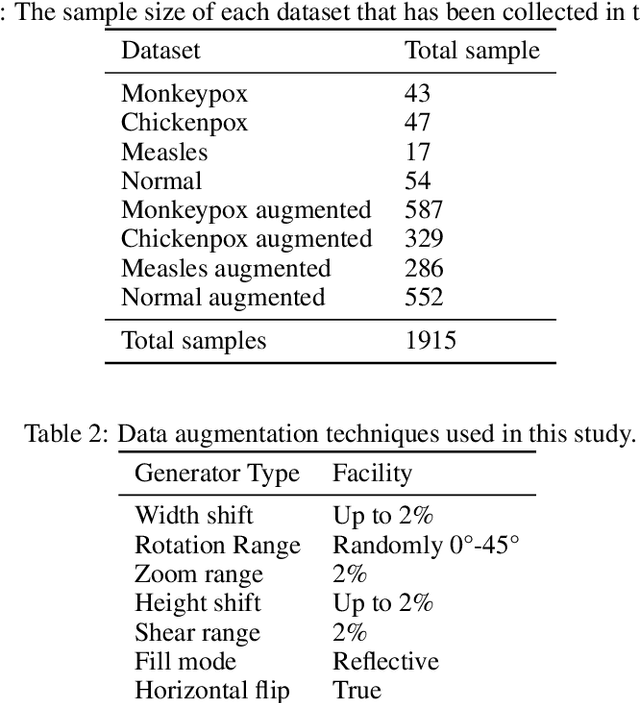
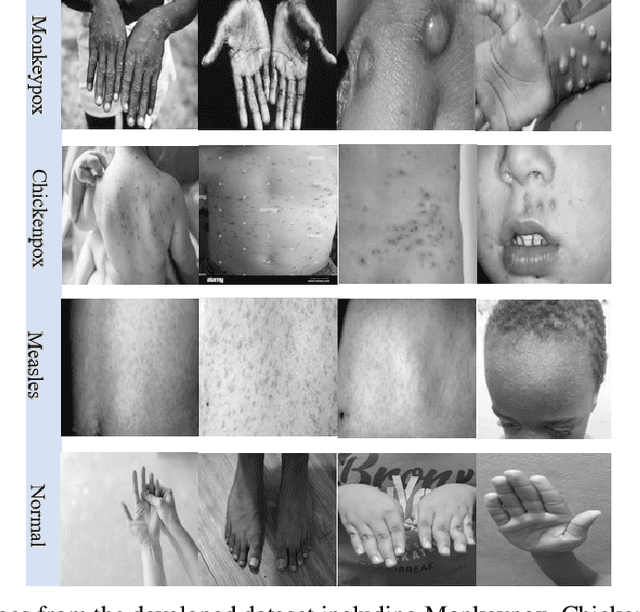
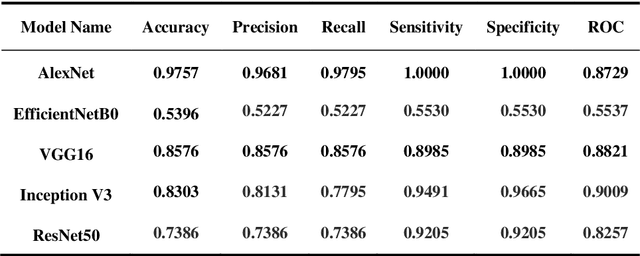
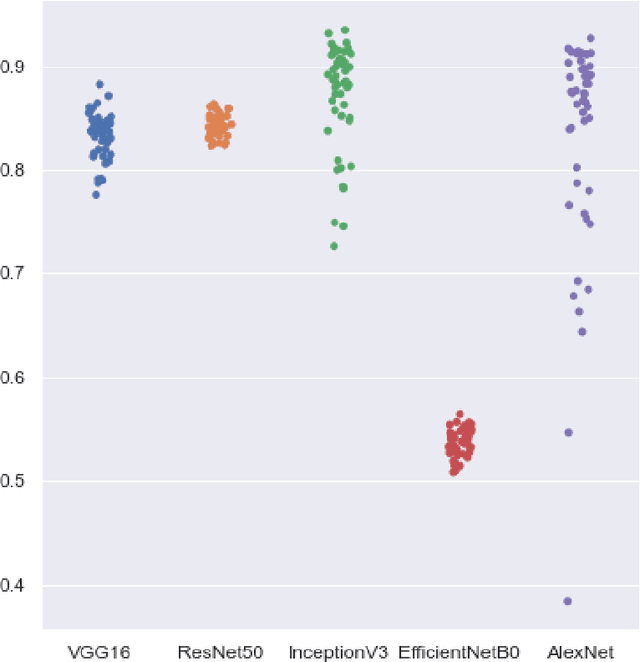
While the world is still attempting to recover from the damage caused by the broad spread of COVID-19, the Monkeypox virus poses a new threat of becoming a global pandemic. Although the Monkeypox virus itself is not deadly and contagious as COVID-19, still every day, new patients case has been reported from many nations. Therefore, it will be no surprise if the world ever faces another global pandemic due to the lack of proper precautious steps. Recently, Machine learning (ML) has demonstrated huge potential in image-based diagnoses such as cancer detection, tumor cell identification, and COVID-19 patient detection. Therefore, a similar application can be adopted to diagnose the Monkeypox-related disease as it infected the human skin, which image can be acquired and further used in diagnosing the disease. Considering this opportunity, in this work, we introduce a newly developed "Monkeypox2022" dataset that is publicly available to use and can be obtained from our shared GitHub repository. The dataset is created by collecting images from multiple open-source and online portals that do not impose any restrictions on use, even for commercial purposes, hence giving a safer path to use and disseminate such data when constructing and deploying any type of ML model. Further, we propose and evaluate a modified VGG16 model, which includes two distinct studies: Study One and Two. Our exploratory computational results indicate that our suggested model can identify Monkeypox patients with an accuracy of $97\pm1.8\%$ (AUC=97.2) and $88\pm0.8\%$ (AUC=0.867) for Study One and Two, respectively. Additionally, we explain our model's prediction and feature extraction utilizing Local Interpretable Model-Agnostic Explanations (LIME) help to a deeper insight into specific features that characterize the onset of the Monkeypox virus.
Oral cancer detection and interpretation: Deep multiple instance learning versus conventional deep single instance learning
Feb 03, 2022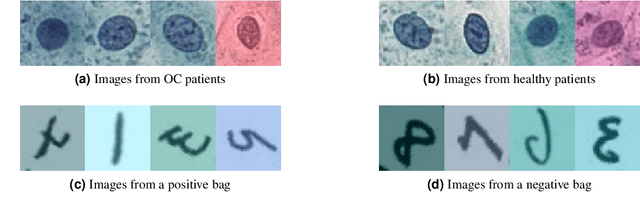
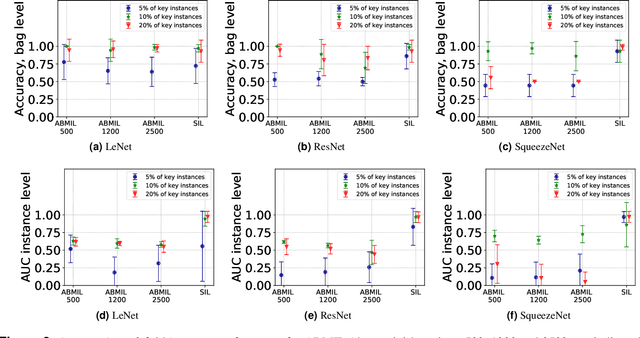
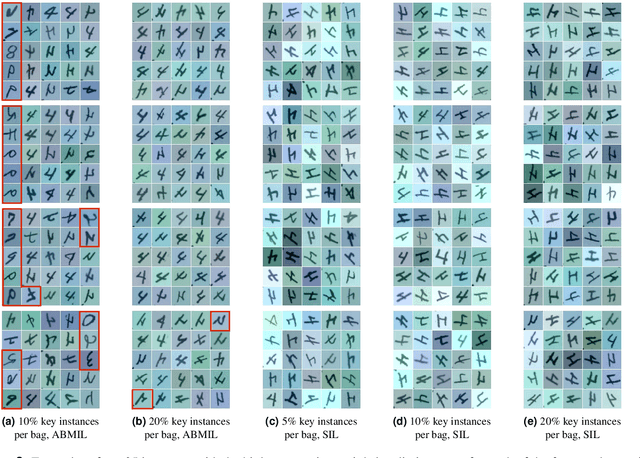
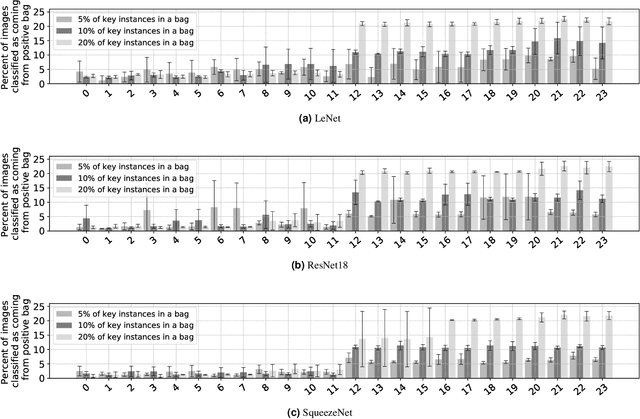
The current medical standard for setting an oral cancer (OC) diagnosis is histological examination of a tissue sample from the oral cavity. This process is time consuming and more invasive than an alternative approach of acquiring a brush sample followed by cytological analysis. Skilled cytotechnologists are able to detect changes due to malignancy, however, to introduce this approach into clinical routine is associated with challenges such as a lack of experts and labour-intensive work. To design a trustworthy OC detection system that would assist cytotechnologists, we are interested in AI-based methods that reliably can detect cancer given only per-patient labels (minimizing annotation bias), and also provide information on which cells are most relevant for the diagnosis (enabling supervision and understanding). We, therefore, perform a comparison of a conventional single instance learning (SIL) approach and a modern multiple instance learning (MIL) method suitable for OC detection and interpretation, utilizing three different neural network architectures. To facilitate systematic evaluation of the considered approaches, we introduce a synthetic PAP-QMNIST dataset, that serves as a model of OC data, while offering access to per-instance ground truth. Our study indicates that on PAP-QMNIST, the SIL performs better, on average, than the MIL approach. Performance at the bag level on real-world cytological data is similar for both methods, yet the single instance approach performs better on average. Visual examination by cytotechnologist indicates that the methods manage to identify cells which deviate from normality, including malignant cells as well as those suspicious for dysplasia. We share the code as open source at https://github.com/MIDA-group/OralCancerMILvsSIL
ProstAttention-Net: A deep attention model for prostate cancer segmentation by aggressiveness in MRI scans
Nov 23, 2022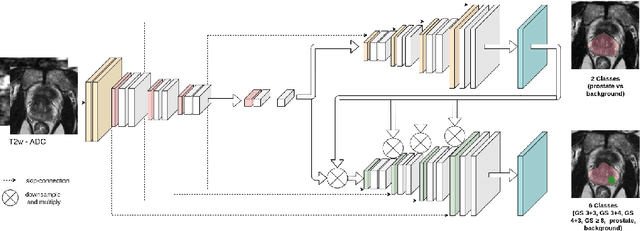
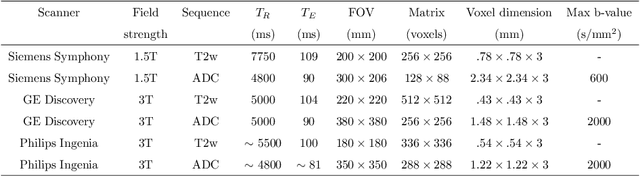
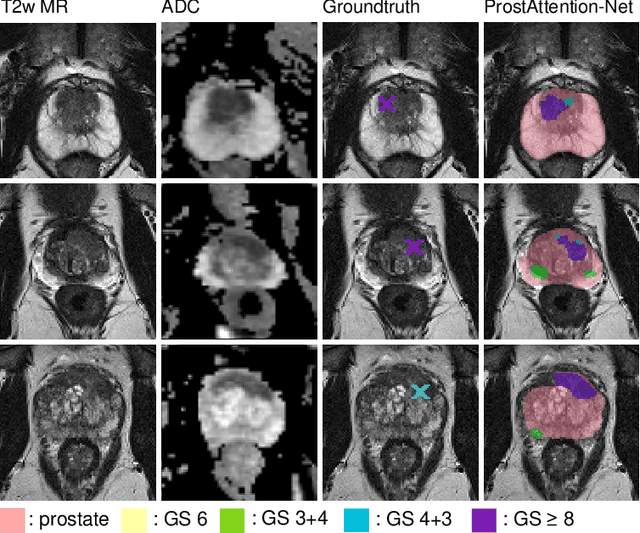
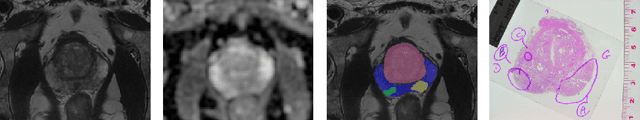
Multiparametric magnetic resonance imaging (mp-MRI) has shown excellent results in the detection of prostate cancer (PCa). However, characterizing prostate lesions aggressiveness in mp-MRI sequences is impossible in clinical practice, and biopsy remains the reference to determine the Gleason score (GS). In this work, we propose a novel end-to-end multi-class network that jointly segments the prostate gland and cancer lesions with GS group grading. After encoding the information on a latent space, the network is separated in two branches: 1) the first branch performs prostate segmentation 2) the second branch uses this zonal prior as an attention gate for the detection and grading of prostate lesions. The model was trained and validated with a 5-fold cross-validation on an heterogeneous series of 219 MRI exams acquired on three different scanners prior prostatectomy. In the free-response receiver operating characteristics (FROC) analysis for clinically significant lesions (defined as GS > 6) detection, our model achieves 69.0% $\pm$14.5% sensitivity at 2.9 false positive per patient on the whole prostate and 70.8% $\pm$14.4% sensitivity at 1.5 false positive when considering the peripheral zone (PZ) only. Regarding the automatic GS group
Identifying Women with Mammographically-Occult Breast Cancer Leveraging GAN-Simulated Mammograms
Sep 24, 2021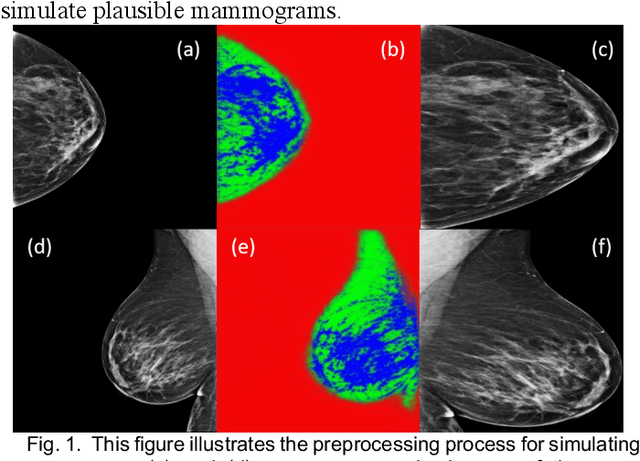
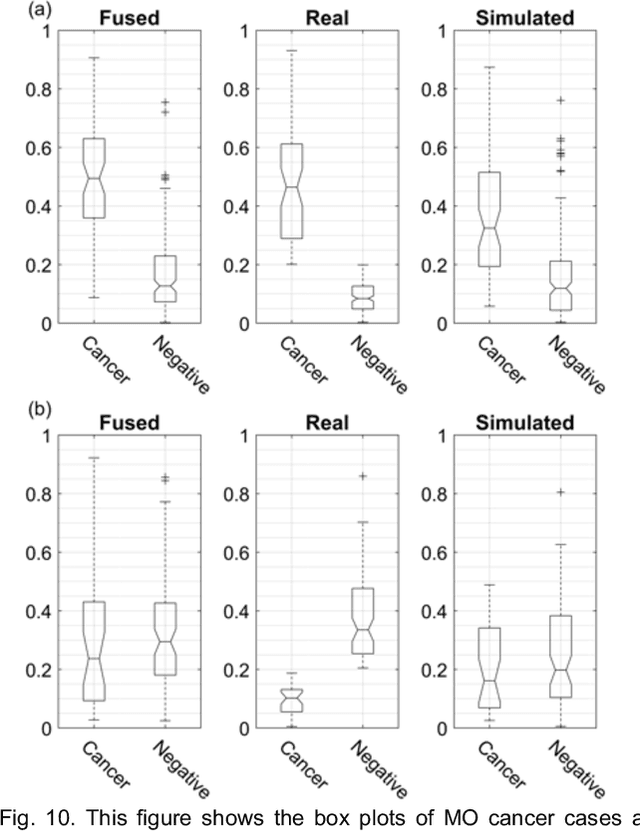
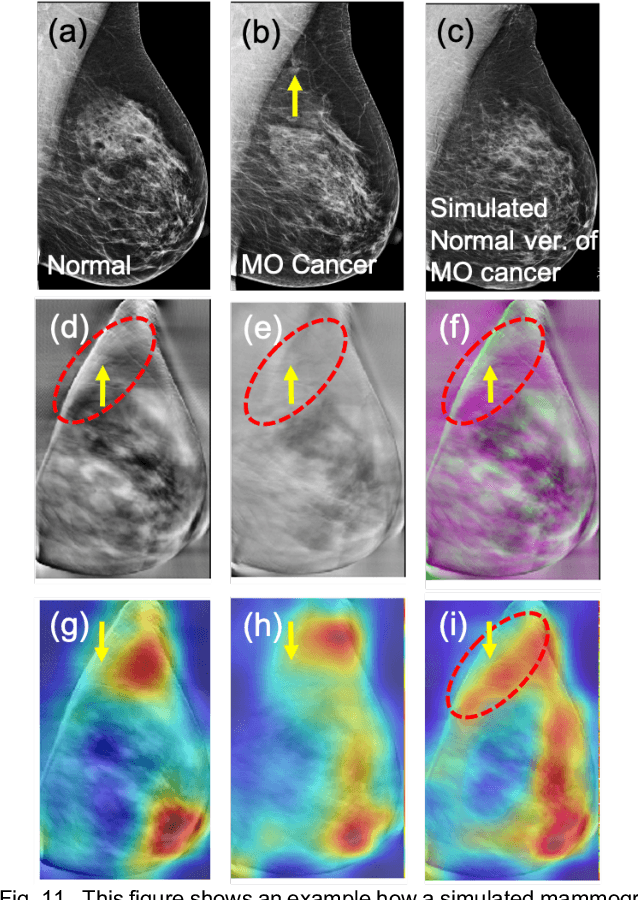
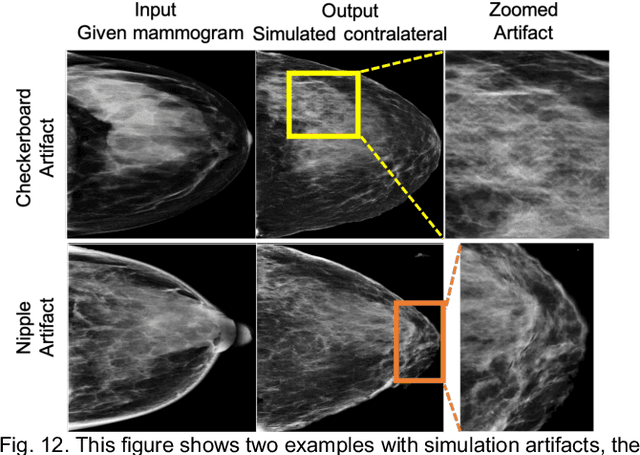
Our objective is to show the feasibility of using simulated mammograms to detect mammographically-occult (MO) cancer in women with dense breasts and a normal screening mammogram who could be triaged for additional screening with magnetic resonance imaging (MRI) or ultrasound. We developed a Conditional Generative Adversarial Network (CGAN) to simulate a mammogram with normal appearance using the opposite mammogram as the condition. We used a Convolutional Neural Network (CNN) trained on Radon Cumulative Distribution Transform (RCDT) processed mammograms to detect MO cancer. For training CGAN, we used screening mammograms of 1366 women. For MO cancer detection, we used screening mammograms of 333 women (97 MO cancer) with dense breasts. We simulated the right mammogram for normal controls and the cancer side for MO cancer cases. We created two RCDT images, one from a real mammogram pair and another from a real-simulated mammogram pair. We finetuned a VGG16 on resulting RCDT images to classify the women with MO cancer. We compared the classification performance of the CNN trained on fused RCDT images, CNN_{Fused} to that of trained only on real RCDT images, CNN_{Real}, and to that of trained only on simulated RCDT images, CNN_{Simulated}. The test AUC for CNN_{Fused} was 0.77 with a 95% confidence interval (95CI) of [0.71, 0.83], which was statistically better (p-value < 0.02) than the CNN_{Real} AUC of 0.70 with a 95CI of [0.64, 0.77] and CNN_{Simulated} AUC of 0.68 with a 95CI of [0.62, 0.75]. It showed that CGAN simulated mammograms can help MO cancer detection.
A Comparative Study of Gastric Histopathology Sub-size Image Classification: from Linear Regression to Visual Transformer
May 25, 2022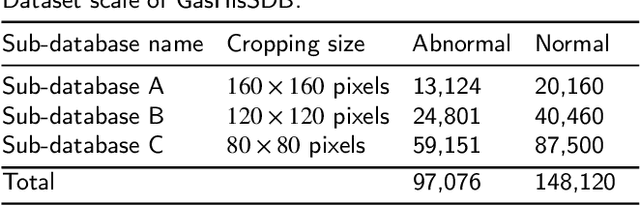
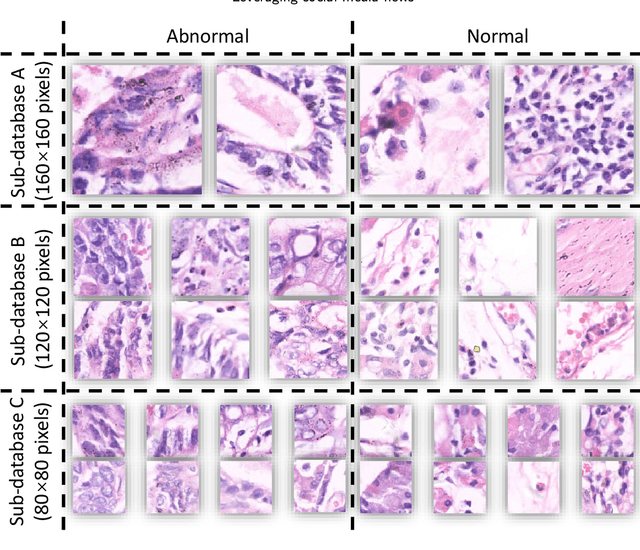
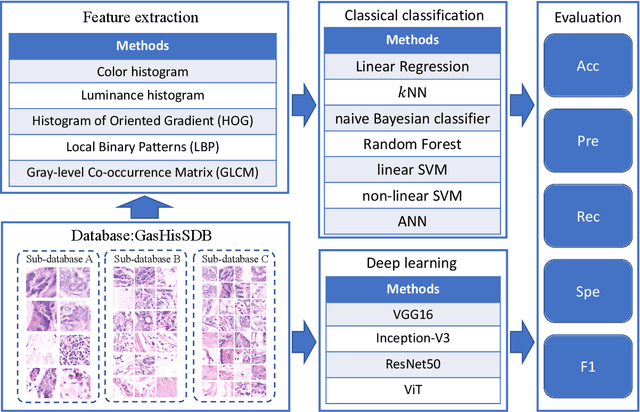
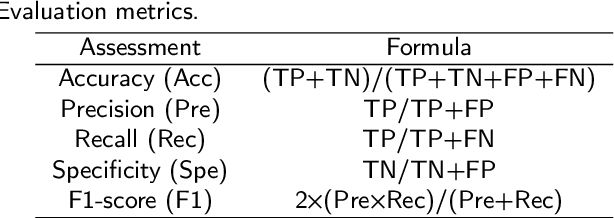
Gastric cancer is the fifth most common cancer in the world. At the same time, it is also the fourth most deadly cancer. Early detection of cancer exists as a guide for the treatment of gastric cancer. Nowadays, computer technology has advanced rapidly to assist physicians in the diagnosis of pathological pictures of gastric cancer. Ensemble learning is a way to improve the accuracy of algorithms, and finding multiple learning models with complementarity types is the basis of ensemble learning. The complementarity of sub-size pathology image classifiers when machine performance is insufficient is explored in this experimental platform. We choose seven classical machine learning classifiers and four deep learning classifiers for classification experiments on the GasHisSDB database. Among them, classical machine learning algorithms extract five different image virtual features to match multiple classifier algorithms. For deep learning, we choose three convolutional neural network classifiers. In addition, we also choose a novel Transformer-based classifier. The experimental platform, in which a large number of classical machine learning and deep learning methods are performed, demonstrates that there are differences in the performance of different classifiers on GasHisSDB. Classical machine learning models exist for classifiers that classify Abnormal categories very well, while classifiers that excel in classifying Normal categories also exist. Deep learning models also exist with multiple models that can be complementarity. Suitable classifiers are selected for ensemble learning, when machine performance is insufficient. This experimental platform demonstrates that multiple classifiers are indeed complementarity and can improve the efficiency of ensemble learning. This can better assist doctors in diagnosis, improve the detection of gastric cancer, and increase the cure rate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge