"cancer detection": models, code, and papers
Scalable Reinforcement-Learning-Based Neural Architecture Search for Cancer Deep Learning Research
Sep 01, 2019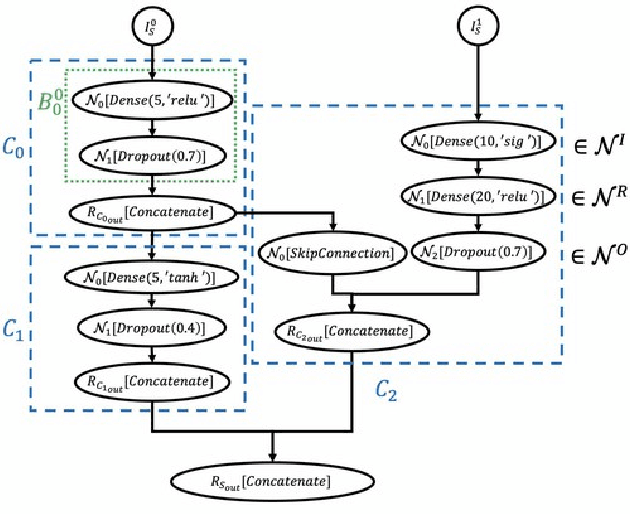
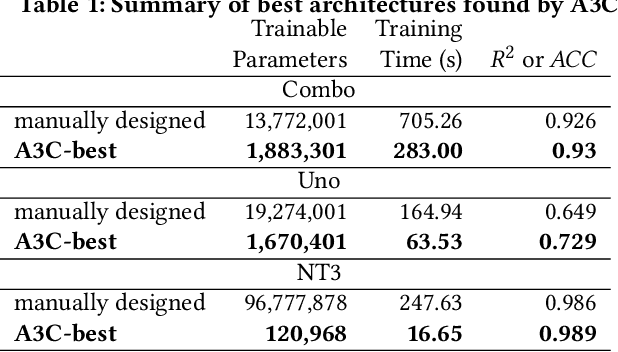
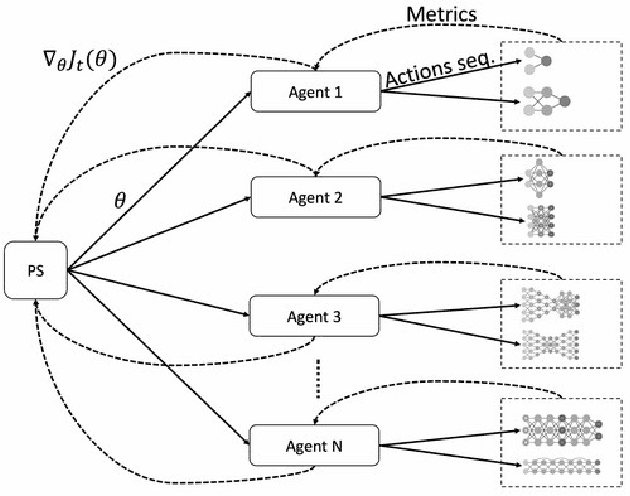
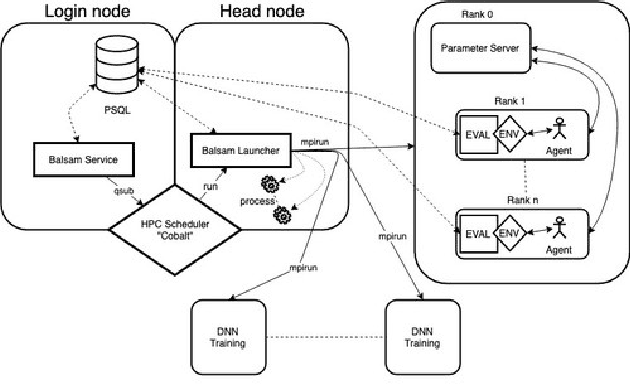
Cancer is a complex disease, the understanding and treatment of which are being aided through increases in the volume of collected data and in the scale of deployed computing power. Consequently, there is a growing need for the development of data-driven and, in particular, deep learning methods for various tasks such as cancer diagnosis, detection, prognosis, and prediction. Despite recent successes, however, designing high-performing deep learning models for nonimage and nontext cancer data is a time-consuming, trial-and-error, manual task that requires both cancer domain and deep learning expertise. To that end, we develop a reinforcement-learning-based neural architecture search to automate deep-learning-based predictive model development for a class of representative cancer data. We develop custom building blocks that allow domain experts to incorporate the cancer-data-specific characteristics. We show that our approach discovers deep neural network architectures that have significantly fewer trainable parameters, shorter training time, and accuracy similar to or higher than those of manually designed architectures. We study and demonstrate the scalability of our approach on up to 1,024 Intel Knights Landing nodes of the Theta supercomputer at the Argonne Leadership Computing Facility.
Interpretative Computer-aided Lung Cancer Diagnosis: from Radiology Analysis to Malignancy Evaluation
Feb 22, 2021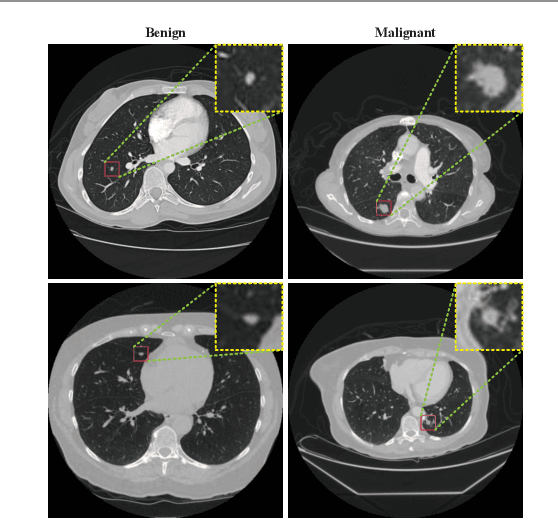
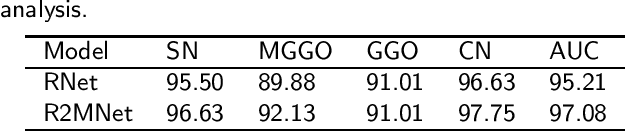
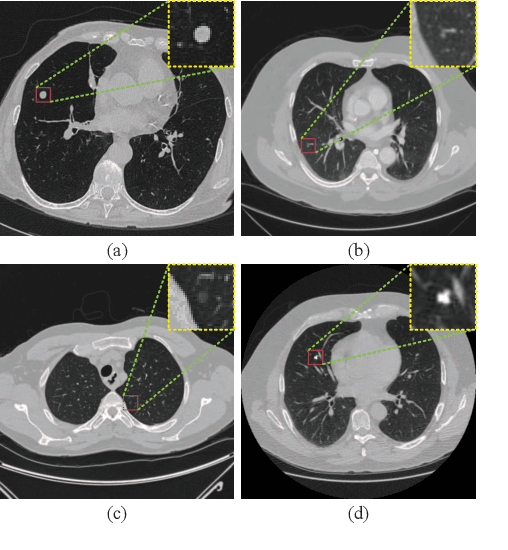
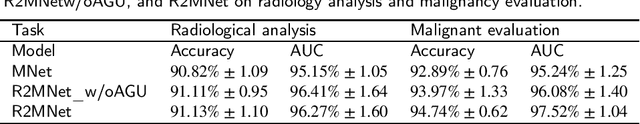
Background and Objective:Computer-aided diagnosis (CAD) systems promote diagnosis effectiveness and alleviate pressure of radiologists. A CAD system for lung cancer diagnosis includes nodule candidate detection and nodule malignancy evaluation. Recently, deep learning-based pulmonary nodule detection has reached satisfactory performance ready for clinical application. However, deep learning-based nodule malignancy evaluation depends on heuristic inference from low-dose computed tomography volume to malignant probability, which lacks clinical cognition. Methods:In this paper, we propose a joint radiology analysis and malignancy evaluation network (R2MNet) to evaluate the pulmonary nodule malignancy via radiology characteristics analysis. Radiological features are extracted as channel descriptor to highlight specific regions of the input volume that are critical for nodule malignancy evaluation. In addition, for model explanations, we propose channel-dependent activation mapping to visualize the features and shed light on the decision process of deep neural network. Results:Experimental results on the LIDC-IDRI dataset demonstrate that the proposed method achieved area under curve of 96.27% on nodule radiology analysis and AUC of 97.52% on nodule malignancy evaluation. In addition, explanations of CDAM features proved that the shape and density of nodule regions were two critical factors that influence a nodule to be inferred as malignant, which conforms with the diagnosis cognition of experienced radiologists. Conclusion:Incorporating radiology analysis with nodule malignant evaluation, the network inference process conforms to the diagnostic procedure of radiologists and increases the confidence of evaluation results. Besides, model interpretation with CDAM features shed light on the regions which DNNs focus on when they estimate nodule malignancy probabilities.
Automatic Generation of Interpretable Lung Cancer Scoring Models from Chest X-Ray Images
Dec 17, 2020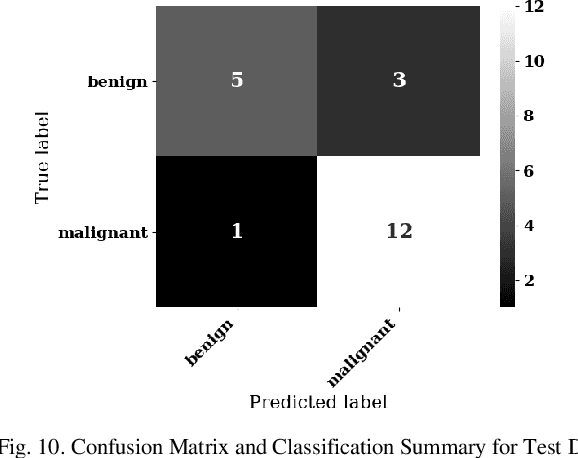
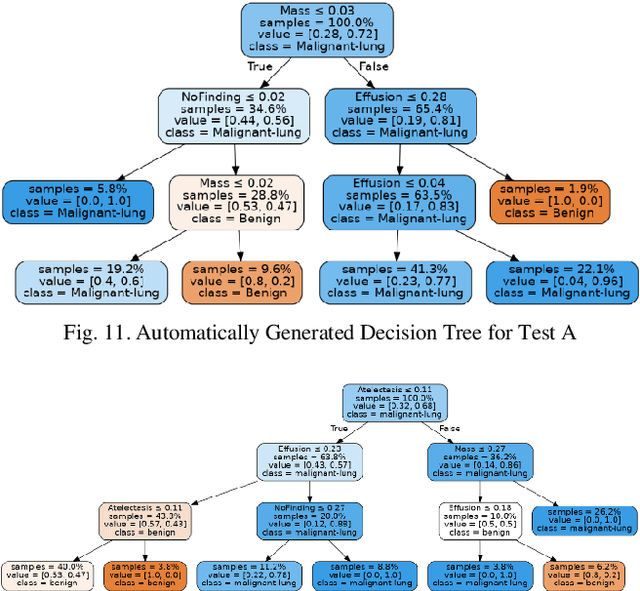
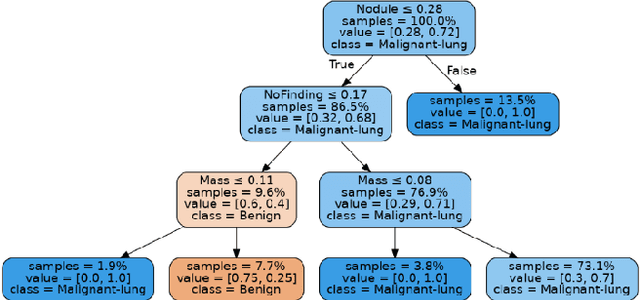
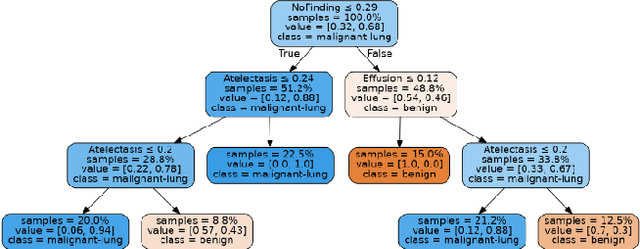
Lung cancer is the leading cause of cancer death worldwide with early detection being the key to a positive patient prognosis. Although a multitude of studies have demonstrated that machine learning, and particularly deep learning, techniques are effective at automatically diagnosing lung cancer, these techniques have yet to be clinically approved and adopted by the medical community. Most research in this field is focused on the narrow task of nodule detection to provide an artificial radiological second reading. We instead focus on extracting, from chest X-ray images, a wider range of pathologies associated with lung cancer using a computer vision model trained on a large dataset. We then find the set of best fit decision trees against an independent, smaller dataset for which lung cancer malignancy metadata is provided. For this small inferencing dataset, our best model achieves sensitivity and specificity of 85% and 75% respectively with a positive predictive value of 85% which is comparable to the performance of human radiologists. Furthermore, the decision trees created by this method may be considered as a starting point for refinement by medical experts into clinically usable multi-variate lung cancer scoring and diagnostic models.
Noncoding RNAs and deep learning neural network discriminate multi-cancer types
Mar 01, 2021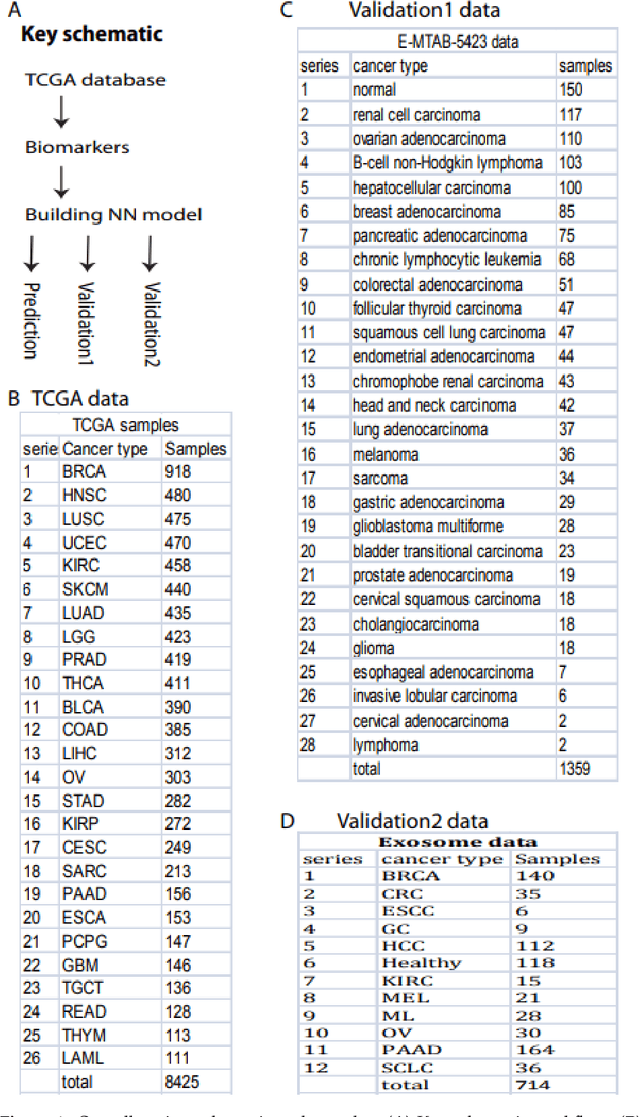
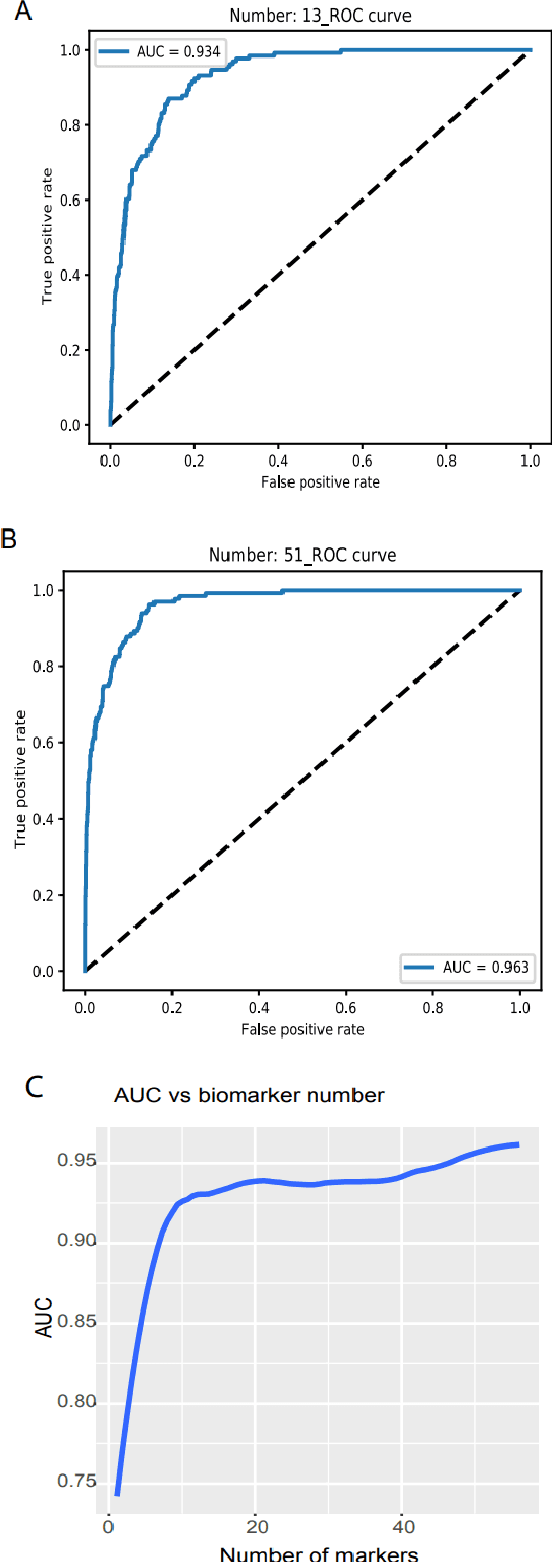
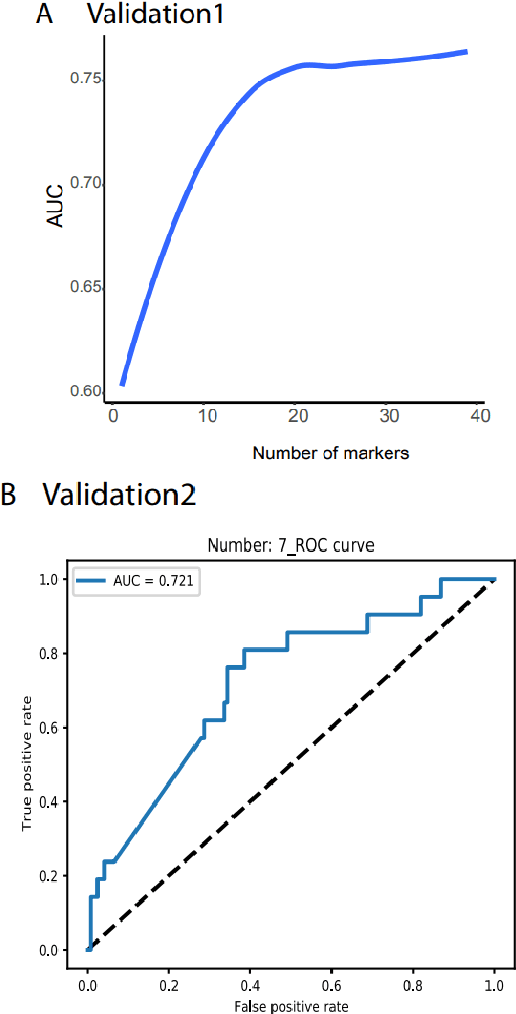
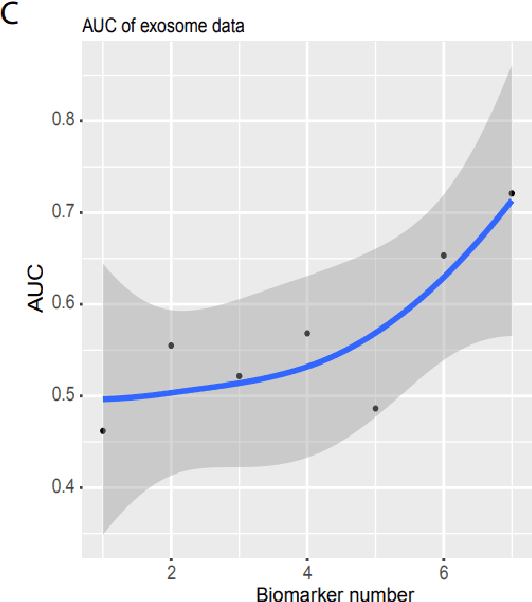
Detecting cancers at early stages can dramatically reduce mortality rates. Therefore, practical cancer screening at the population level is needed. Here, we develop a comprehensive detection system to classify all common cancer types. By integrating artificial intelligence deep learning neural network and noncoding RNA biomarkers selected from massive data, our system can accurately detect cancer vs healthy object with 96.3% of AUC of ROC (Area Under Curve of a Receiver Operating Characteristic curve). Intriguinely, with no more than 6 biomarkers, our approach can easily discriminate any individual cancer type vs normal with 99% to 100% AUC. Furthermore, a comprehensive marker panel can simultaneously multi-classify all common cancers with a stable 78% of accuracy at heterological cancerous tissues and conditions. This provides a valuable framework for large scale cancer screening. The AI models and plots of results were available in https://combai.org/ai/cancerdetection/
Method and System for Image Analysis to Detect Cancer
Aug 26, 2019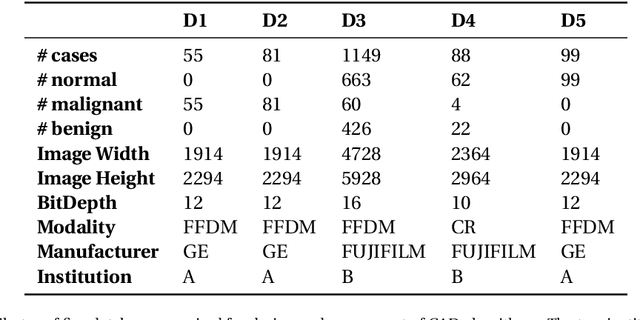
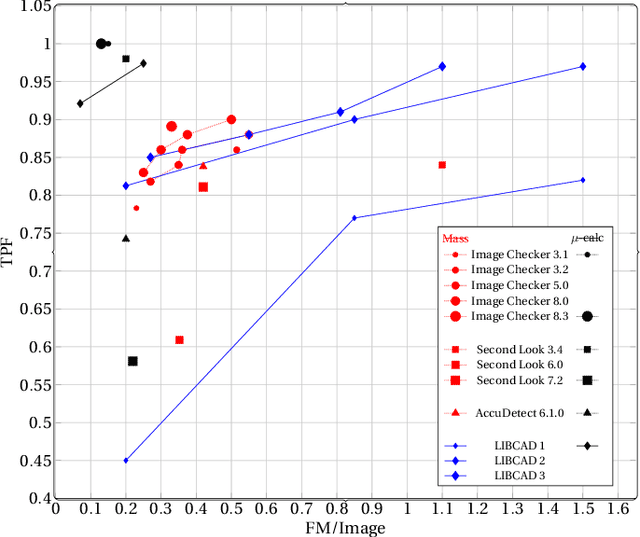
Breast cancer is the most common cancer and is the leading cause of cancer death among women worldwide. Detection of breast cancer, while it is still small and confined to the breast, provides the best chance of effective treatment. Computer Aided Detection (CAD) systems that detect cancer from mammograms will help in reducing the human errors that lead to missing breast carcinoma. Literature is rich of scientific papers for methods of CAD design, yet with no complete system architecture to deploy those methods. On the other hand, commercial CADs are developed and deployed only to vendors' mammography machines with no availability to public access. This paper presents a complete CAD; it is complete since it combines, on a hand, the rigor of algorithm design and assessment (method), and, on the other hand, the implementation and deployment of a system architecture for public accessibility (system). (1) We develop a novel algorithm for image enhancement so that mammograms acquired from any digital mammography machine look qualitatively of the same clarity to radiologists' inspection; and is quantitatively standardized for the detection algorithms. (2) We develop novel algorithms for masses and microcalcifications detection with accuracy superior to both literature results and the majority of approved commercial systems. (3) We design, implement, and deploy a system architecture that is computationally effective to allow for deploying these algorithms to cloud for public access.
DeepCervix: A Deep Learning-based Framework for the Classification of Cervical Cells Using Hybrid Deep Feature Fusion Techniques
Feb 24, 2021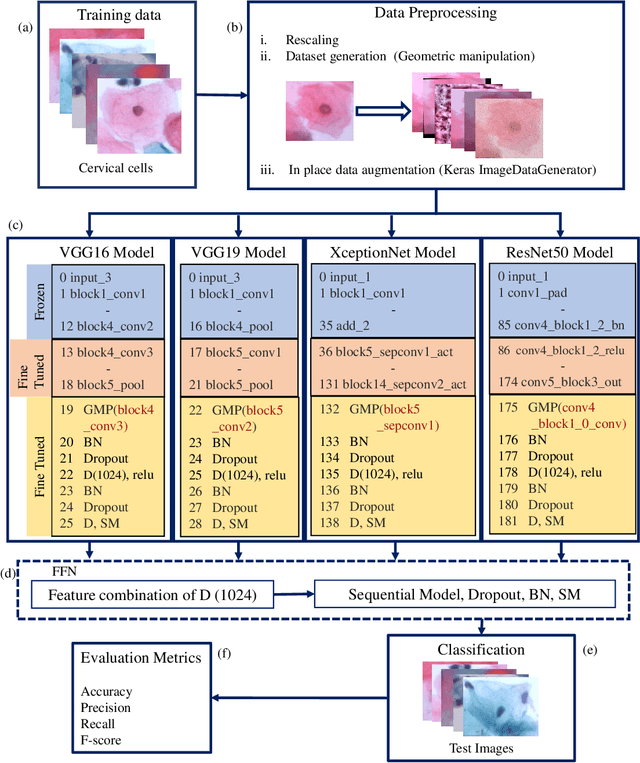
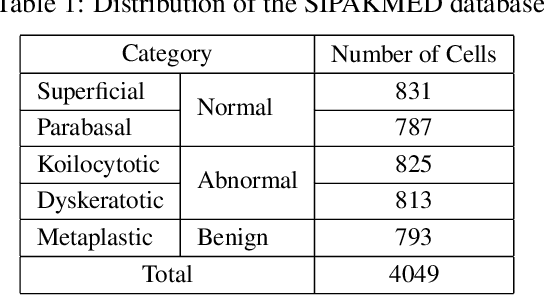
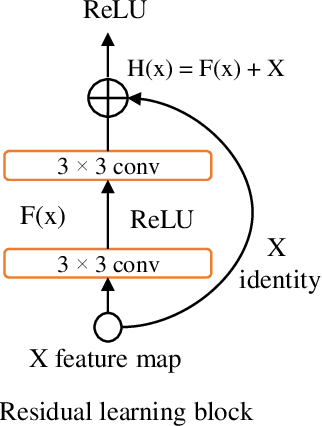
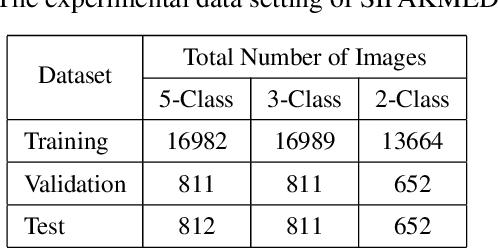
Cervical cancer, one of the most common fatal cancers among women, can be prevented by regular screening to detect any precancerous lesions at early stages and treat them. Pap smear test is a widely performed screening technique for early detection of cervical cancer, whereas this manual screening method suffers from high false-positive results because of human errors. To improve the manual screening practice, machine learning (ML) and deep learning (DL) based computer-aided diagnostic (CAD) systems have been investigated widely to classify cervical pap cells. Most of the existing researches require pre-segmented images to obtain good classification results, whereas accurate cervical cell segmentation is challenging because of cell clustering. Some studies rely on handcrafted features, which cannot guarantee the classification stage's optimality. Moreover, DL provides poor performance for a multiclass classification task when there is an uneven distribution of data, which is prevalent in the cervical cell dataset. This investigation has addressed those limitations by proposing DeepCervix, a hybrid deep feature fusion (HDFF) technique based on DL to classify the cervical cells accurately. Our proposed method uses various DL models to capture more potential information to enhance classification performance. Our proposed HDFF method is tested on the publicly available SIPAKMED dataset and compared the performance with base DL models and the LF method. For the SIPAKMED dataset, we have obtained the state-of-the-art classification accuracy of 99.85%, 99.38%, and 99.14% for 2-class, 3-class, and 5-class classification. Moreover, our method is tested on the Herlev dataset and achieves an accuracy of 98.32% for binary class and 90.32% for 7-class classification.
Improving prostate whole gland segmentation in t2-weighted MRI with synthetically generated data
Mar 27, 2021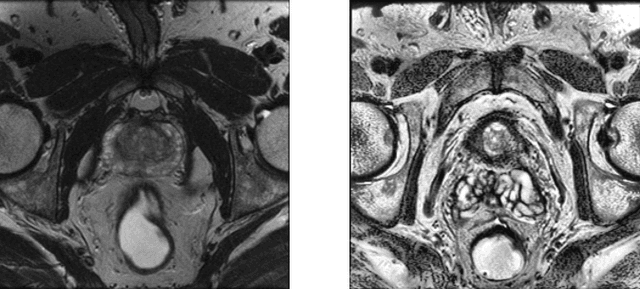
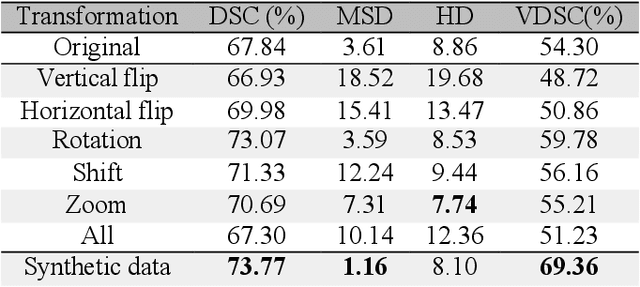
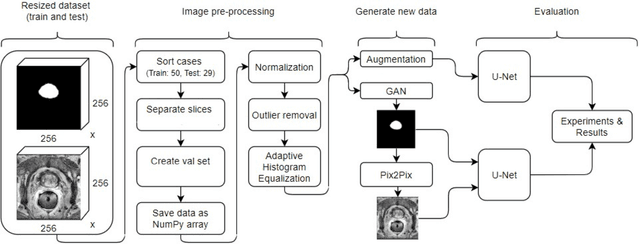
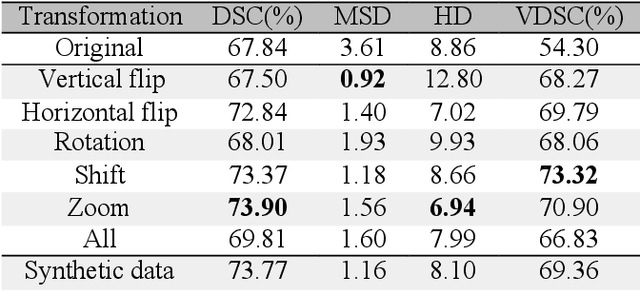
Whole gland (WG) segmentation of the prostate plays a crucial role in detection, staging and treatment planning of prostate cancer (PCa). Despite promise shown by deep learning (DL) methods, they rely on the availability of a considerable amount of annotated data. Augmentation techniques such as translation and rotation of images present an alternative to increase data availability. Nevertheless, the amount of information provided by the transformed data is limited due to the correlation between the generated data and the original. Based on the recent success of generative adversarial networks (GAN) in producing synthetic images for other domains as well as in the medical domain, we present a pipeline to generate WG segmentation masks and synthesize T2-weighted MRI of the prostate based on a publicly available multi-center dataset. Following, we use the generated data as a form of data augmentation. Results show an improvement in the quality of the WG segmentation when compared to standard augmentation techniques.
Handling uncertainty using features from pathology: opportunities in primary care data for developing high risk cancer survival methods
Dec 17, 2020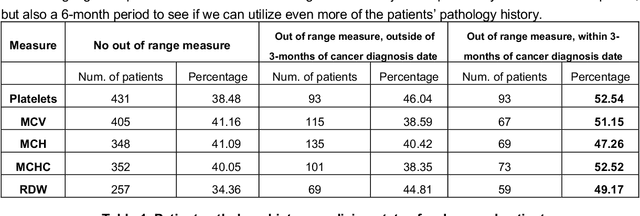
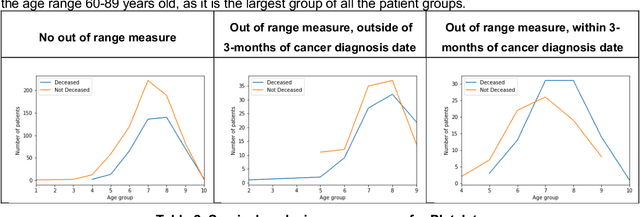
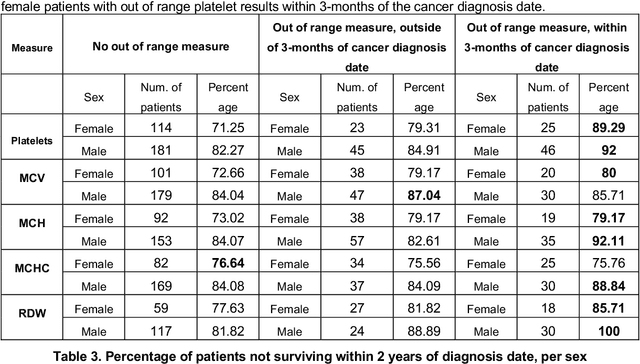
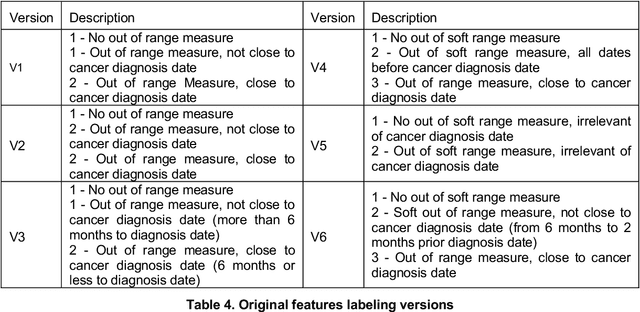
More than 144 000 Australians were diagnosed with cancer in 2019. The majority will first present to their GP symptomatically, even for cancer for which screening programs exist. Diagnosing cancer in primary care is challenging due to the non-specific nature of cancer symptoms and its low prevalence. Understanding the epidemiology of cancer symptoms and patterns of presentation in patient's medical history from primary care data could be important to improve earlier detection and cancer outcomes. As past medical data about a patient can be incomplete, irregular or missing, this creates additional challenges when attempting to use the patient's history for any new diagnosis. Our research aims to investigate the opportunities in a patient's pathology history available to a GP, initially focused on the results within the frequently ordered full blood count to determine relevance to a future high-risk cancer prognosis, and treatment outcome. We investigated how past pathology test results can lead to deriving features that can be used to predict cancer outcomes, with emphasis on patients at risk of not surviving the cancer within 2-year period. This initial work focuses on patients with lung cancer, although the methodology can be applied to other types of cancer and other data within the medical record. Our findings indicate that even in cases of incomplete or obscure patient history, hematological measures can be useful in generating features relevant for predicting cancer risk and survival. The results strongly indicate to add the use of pathology test data for potential high-risk cancer diagnosis, and the utilize additional pathology metrics or other primary care datasets even more for similar purposes.
Automatic Lesion Detection System (ALDS) for Skin Cancer Classification Using SVM and Neural Classifiers
Mar 13, 2020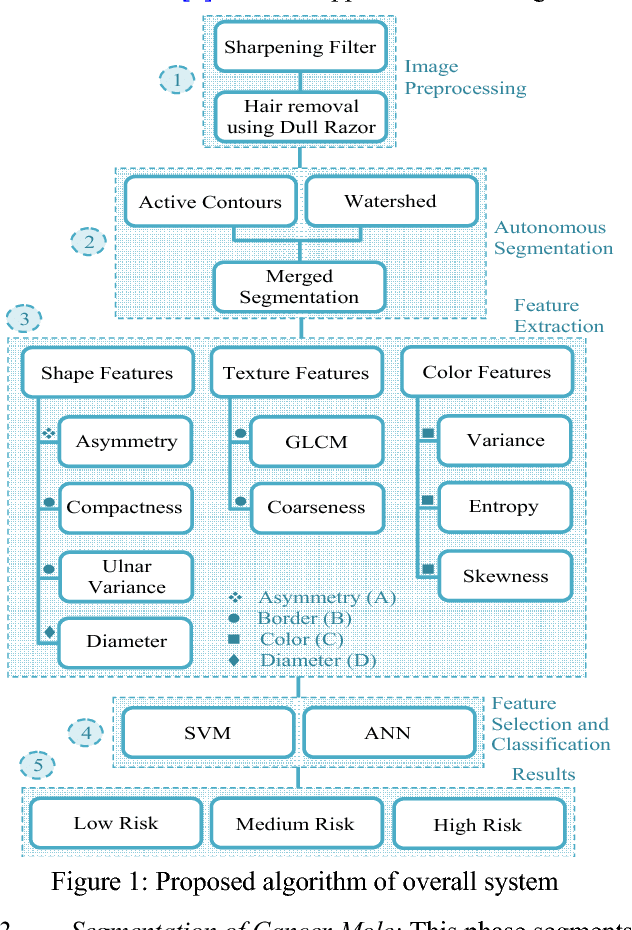

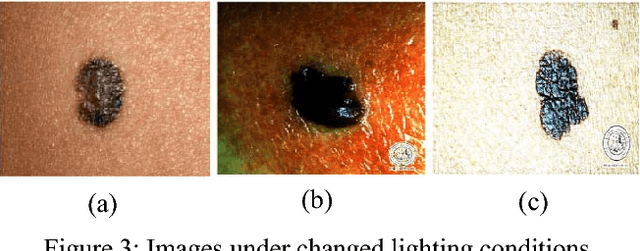
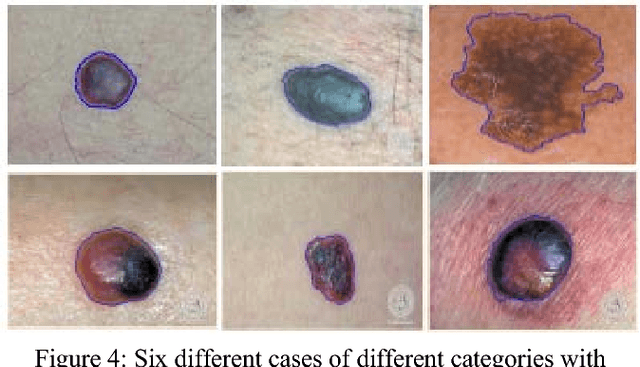
Technology aided platforms provide reliable tools in almost every field these days. These tools being supported by computational power are significant for applications that need sensitive and precise data analysis. One such important application in the medical field is Automatic Lesion Detection System (ALDS) for skin cancer classification. Computer aided diagnosis helps physicians and dermatologists to obtain a second opinion for proper analysis and treatment of skin cancer. Precise segmentation of the cancerous mole along with surrounding area is essential for proper analysis and diagnosis. This paper is focused towards the development of improved ALDS framework based on probabilistic approach that initially utilizes active contours and watershed merged mask for segmenting out the mole and later SVM and Neural Classifier are applied for the classification of the segmented mole. After lesion segmentation, the selected features are classified to ascertain that whether the case under consideration is melanoma or non-melanoma. The approach is tested for varying datasets and comparative analysis is performed that reflects the effectiveness of the proposed system.
Mitosis Detection Under Limited Annotation: A Joint Learning Approach
Jul 02, 2020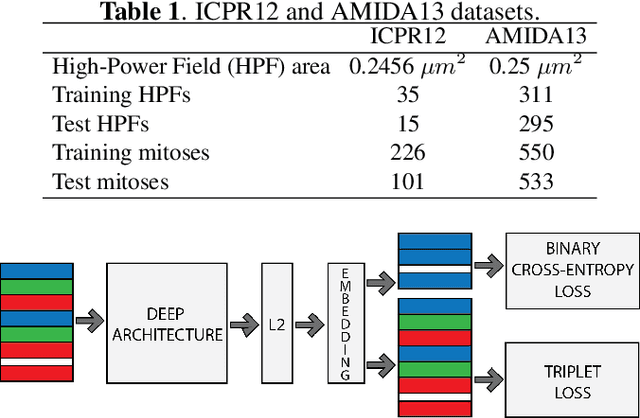
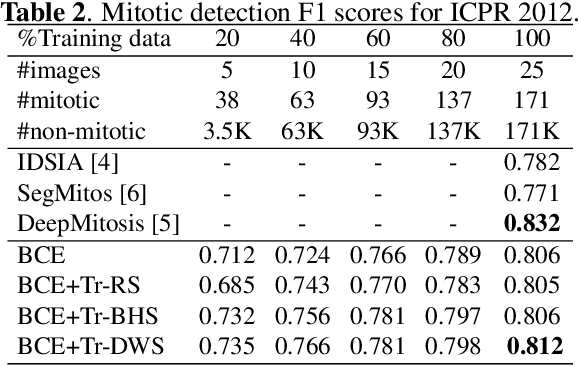
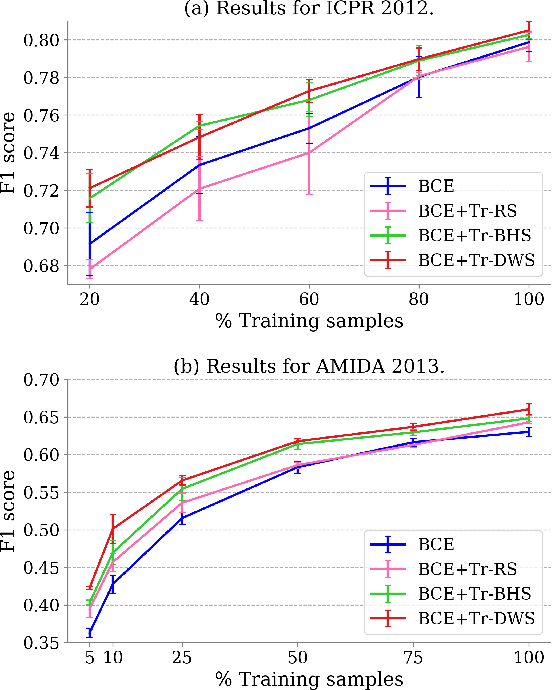
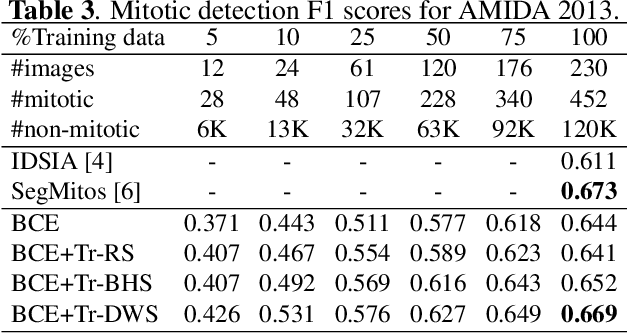
Mitotic counting is a vital prognostic marker of tumor proliferation in breast cancer. Deep learning-based mitotic detection is on par with pathologists, but it requires large labeled data for training. We propose a deep classification framework for enhancing mitosis detection by leveraging class label information, via softmax loss, and spatial distribution information among samples, via distance metric learning. We also investigate strategies towards steadily providing informative samples to boost the learning. The efficacy of the proposed framework is established through evaluation on ICPR 2012 and AMIDA 2013 mitotic data. Our framework significantly improves the detection with small training data and achieves on par or superior performance compared to state-of-the-art methods for using the entire training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge