"cancer detection": models, code, and papers
A Precision Diagnostic Framework of Renal Cell Carcinoma on Whole-Slide Images using Deep Learning
Oct 26, 2021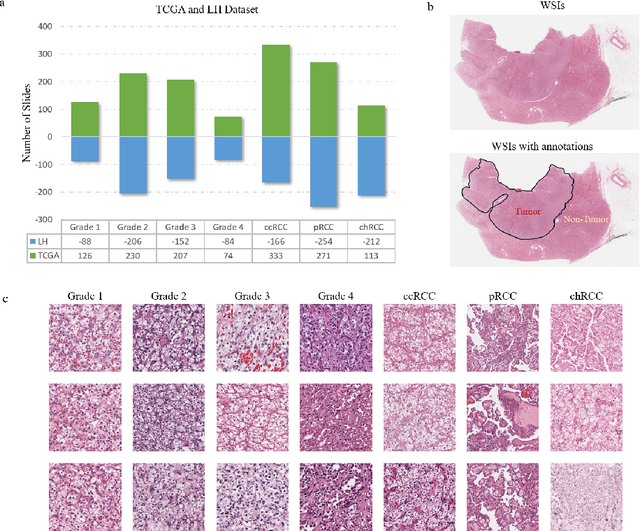
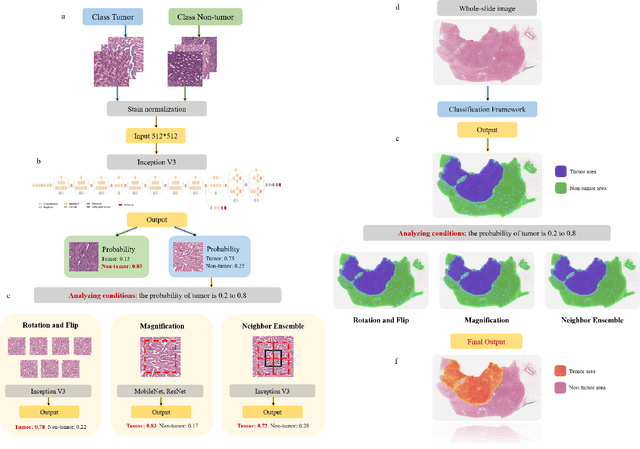
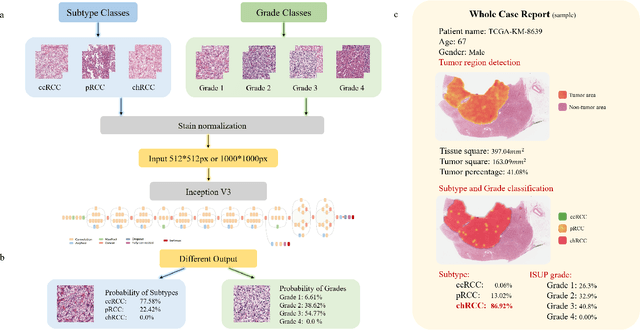
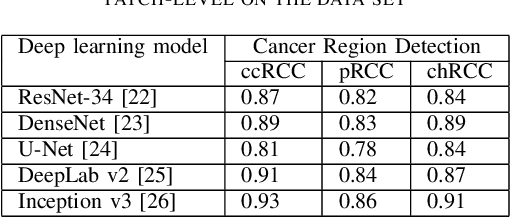
Diagnostic pathology, which is the basis and gold standard of cancer diagnosis, provides essential information on the prognosis of the disease and vital evidence for clinical treatment. Tumor region detection, subtype and grade classification are the fundamental diagnostic indicators for renal cell carcinoma (RCC) in whole-slide images (WSIs). However, pathological diagnosis is subjective, differences in observation and diagnosis between pathologists is common in hospitals with inadequate diagnostic capacity. The main challenge for developing deep learning based RCC diagnostic system is the lack of large-scale datasets with precise annotations. In this work, we proposed a deep learning-based framework for analyzing histopathological images of patients with renal cell carcinoma, which has the potential to achieve pathologist-level accuracy in diagnosis. A deep convolutional neural network (InceptionV3) was trained on the high-quality annotated dataset of The Cancer Genome Atlas (TCGA) whole-slide histopathological image for accurate tumor area detection, classification of RCC subtypes, and ISUP grades classification of clear cell carcinoma subtypes. These results suggest that our framework can help pathologists in the detection of cancer region and classification of subtypes and grades, which could be applied to any cancer type, providing auxiliary diagnosis and promoting clinical consensus.
A Pragmatic Machine Learning Approach to Quantify Tumor Infiltrating Lymphocytes in Whole Slide Images
Feb 14, 2022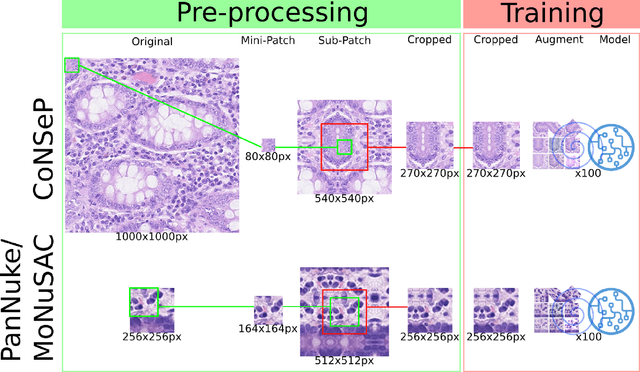
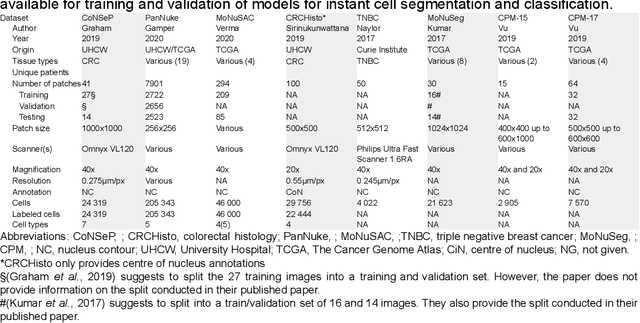
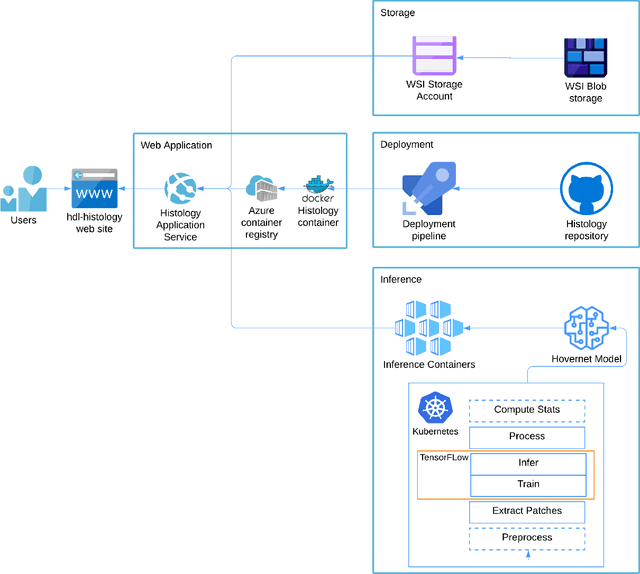
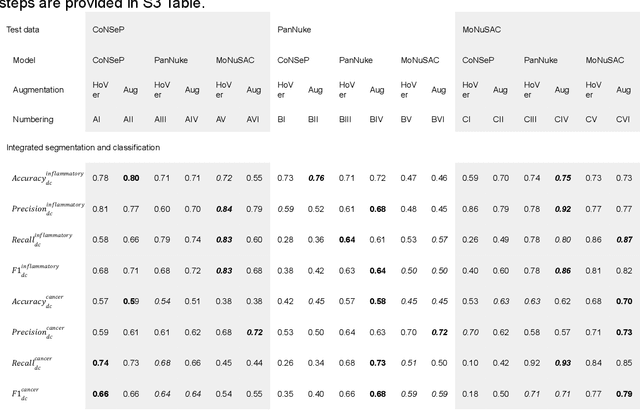
Increased levels of tumor infiltrating lymphocytes (TILs) in cancer tissue indicate favourable outcomes in many types of cancer. Manual quantification of immune cells is inaccurate and time consuming for pathologists. Our aim is to leverage a computational solution to automatically quantify TILs in whole slide images (WSIs) of standard diagnostic haematoxylin and eosin stained sections (H&E slides) from lung cancer patients. Our approach is to transfer an open source machine learning method for segmentation and classification of nuclei in H&E slides trained on public data to TIL quantification without manual labeling of our data. Our results show that additional augmentation improves model transferability when training on few samples/limited tissue types. Models trained with sufficient samples/tissue types do not benefit from our additional augmentation policy. Further, the resulting TIL quantification correlates to patient prognosis and compares favorably to the current state-of-the-art method for immune cell detection in non-small lung cancer (current standard CD8 cells in DAB stained TMAs HR 0.34 95% CI 0.17-0.68 vs TILs in HE WSIs: HoVer-Net PanNuke Aug Model HR 0.30 95% CI 0.15-0.60, HoVer-Net MoNuSAC Aug model HR 0.27 95% CI 0.14-0.53). Moreover, we implemented a cloud based system to train, deploy and visually inspect machine learning based annotation for H&E slides. Our pragmatic approach bridges the gap between machine learning research, translational clinical research and clinical implementation. However, validation in prospective studies is needed to assert that the method works in a clinical setting.
Whole-Slide Image Focus Quality: Automatic Assessment and Impact on AI Cancer Detection
Jan 15, 2019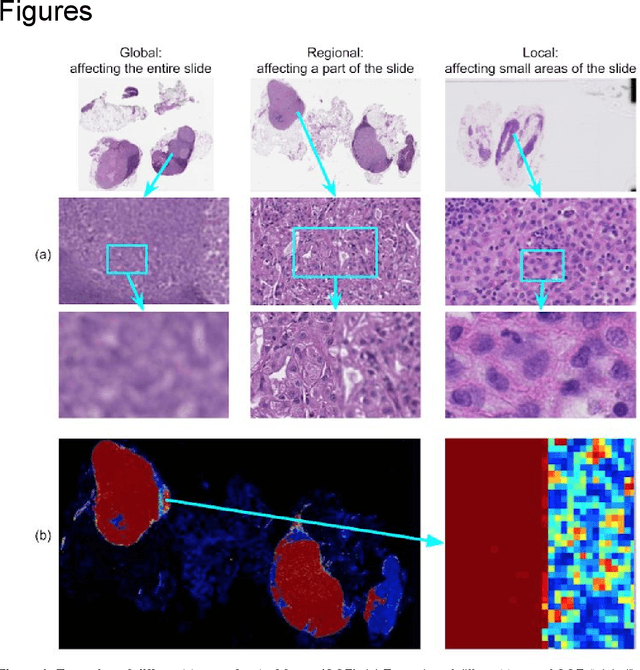
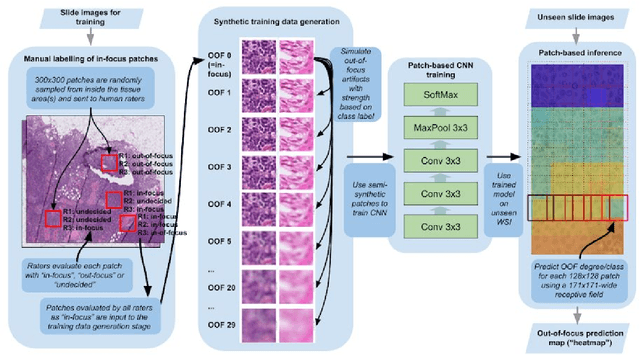
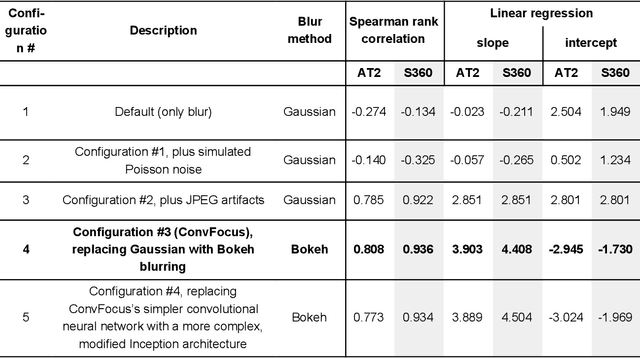
Digital pathology enables remote access or consults and powerful image analysis algorithms. However, the slide digitization process can create artifacts such as out-of-focus (OOF). OOF is often only detected upon careful review, potentially causing rescanning and workflow delays. Although scan-time operator screening for whole-slide OOF is feasible, manual screening for OOF affecting only parts of a slide is impractical. We developed a convolutional neural network (ConvFocus) to exhaustively localize and quantify the severity of OOF regions on digitized slides. ConvFocus was developed using our refined semi-synthetic OOF data generation process, and evaluated using real whole-slide images spanning 3 different tissue types and 3 different stain types that were digitized by two different scanners. ConvFocus's predictions were compared with pathologist-annotated focus quality grades across 514 distinct regions representing 37,700 35x35{\mu}m image patches, and 21 digitized "z-stack" whole-slide images that contain known OOF patterns. When compared to pathologist-graded focus quality, ConvFocus achieved Spearman rank coefficients of 0.81 and 0.94 on two scanners, and reproduced the expected OOF patterns from z-stack scanning. We also evaluated the impact of OOF on the accuracy of a state-of-the-art metastatic breast cancer detector and saw a consistent decrease in performance with increasing OOF. Comprehensive whole-slide OOF categorization could enable rescans prior to pathologist review, potentially reducing the impact of digitization focus issues on the clinical workflow. We show that the algorithm trained on our semi-synthetic OOF data generalizes well to real OOF regions across tissue types, stains, and scanners. Finally, quantitative OOF maps can flag regions that might otherwise be misclassified by image analysis algorithms, preventing OOF-induced errors.
Unsupervised Prostate Cancer Detection on H&E using Convolutional Adversarial Autoencoders
Apr 19, 2018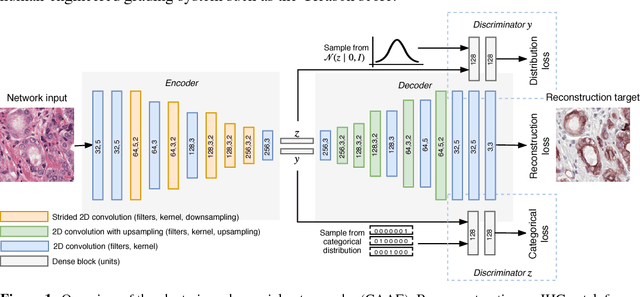
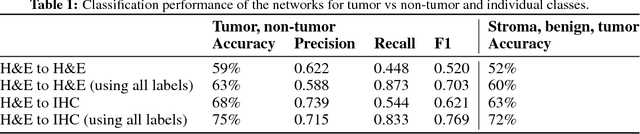
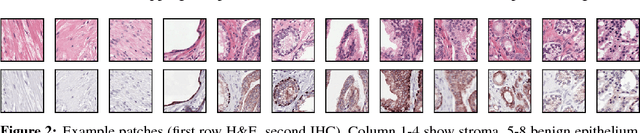
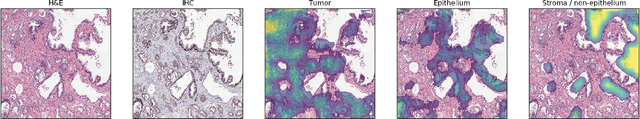
We propose an unsupervised method using self-clustering convolutional adversarial autoencoders to classify prostate tissue as tumor or non-tumor without any labeled training data. The clustering method is integrated into the training of the autoencoder and requires only little post-processing. Our network trains on hematoxylin and eosin (H&E) input patches and we tested two different reconstruction targets, H&E and immunohistochemistry (IHC). We show that antibody-driven feature learning using IHC helps the network to learn relevant features for the clustering task. Our network achieves a F1 score of 0.62 using only a small set of validation labels to assign classes to clusters.
Mitosis Detection for Breast Cancer Pathology Images using UV-Net
Sep 21, 2021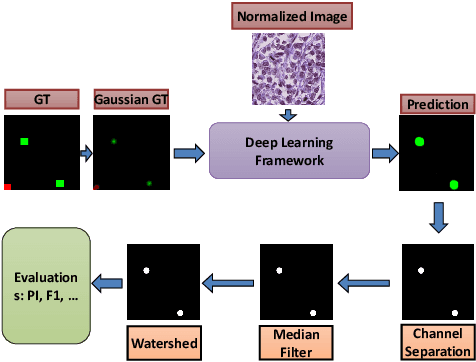
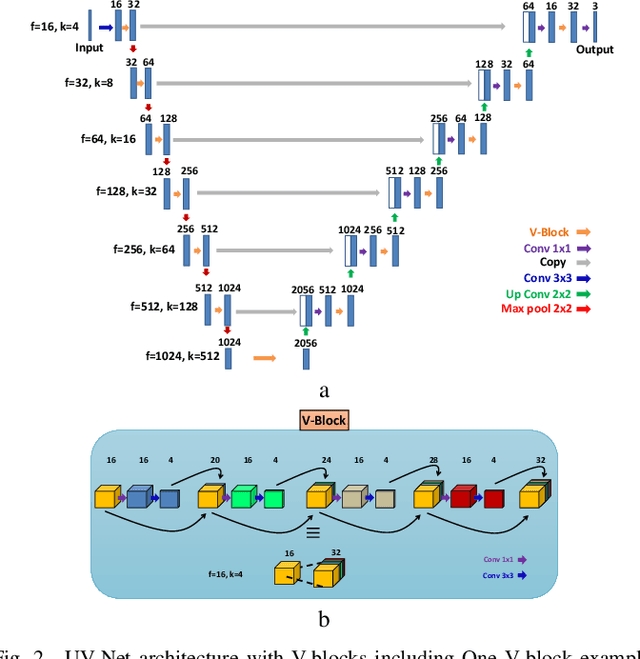
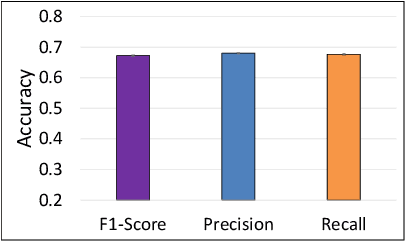
The difficulty of detecting mitosis and its similarity to non-mitosis objects has remained a challenge in computational pathology. The lack of publicly available data has added more complexity. Deep learning algorithms have shown potentials in mitosis detection tasks. However, they face challenges when applied to pathology images with dense medium and diverse dataset. This paper introduces an optimized UV-Net architecture, developed to focus on mitosis details with high-resolution through feature preservation. Stain normalization methods are used to generalize the trained network. An F1 score of 0.6721 is achieved using this network.
Deep Learning-based mitosis detection in breast cancer histologic samples
Sep 02, 2021This is the submission for mitosis detection in the context of the MIDOG 2021 challenge. It is based on the two-stage objection model Faster RCNN as well as DenseNet as a backbone for the neural network architecture. It achieves a F1-score of 0.6645 on the Preliminary Test Phase Leaderboard.
Validation of a deep learning mammography model in a population with low screening rates
Nov 01, 2019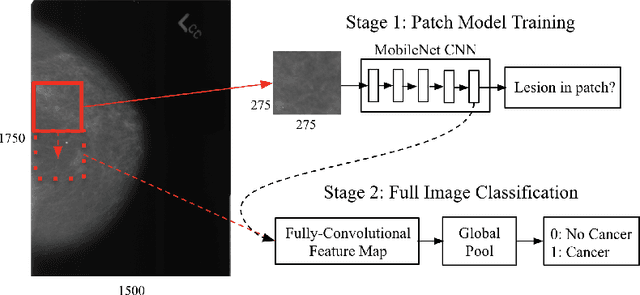
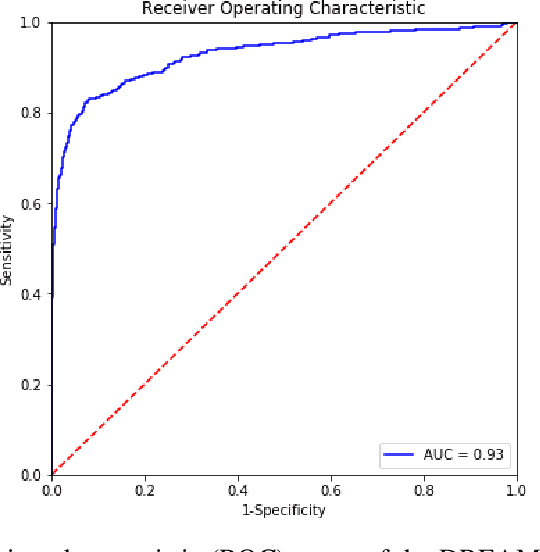
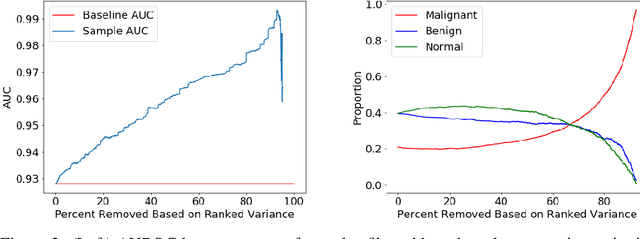
A key promise of AI applications in healthcare is in increasing access to quality medical care in under-served populations and emerging markets. However, deep learning models are often only trained on data from advantaged populations that have the infrastructure and resources required for large-scale data collection. In this paper, we aim to empirically investigate the potential impact of such biases on breast cancer detection in mammograms. We specifically explore how a deep learning algorithm trained on screening mammograms from the US and UK generalizes to mammograms collected at a hospital in China, where screening is not widely implemented. For the evaluation, we use a top-scoring model developed for the Digital Mammography DREAM Challenge. Despite the change in institution and population composition, we find that the model generalizes well, exhibiting similar performance to that achieved in the DREAM Challenge, even when controlling for tumor size. We also illustrate a simple but effective method for filtering predictions based on model variance, which can be particularly useful for deployment in new settings. While there are many components in developing a clinically effective system, these results represent a promising step towards increasing access to life-saving screening mammography in populations where screening rates are currently low.
Distill-to-Label: Weakly Supervised Instance Labeling Using Knowledge Distillation
Jul 26, 2019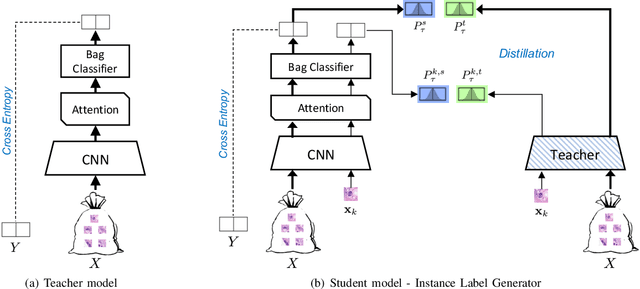
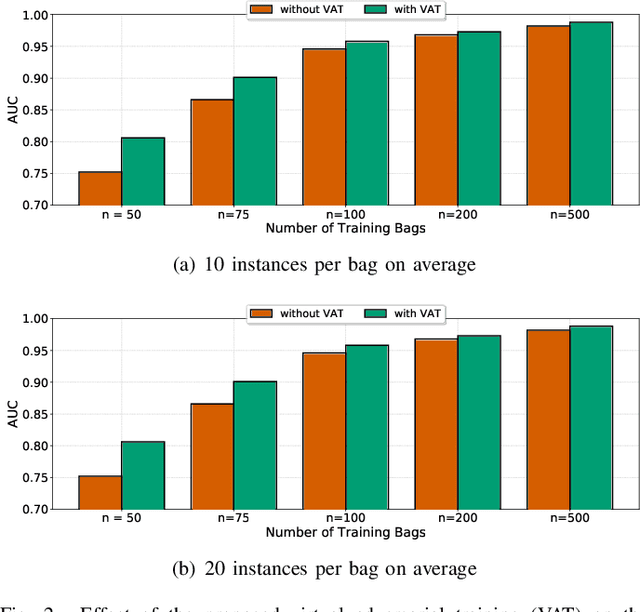
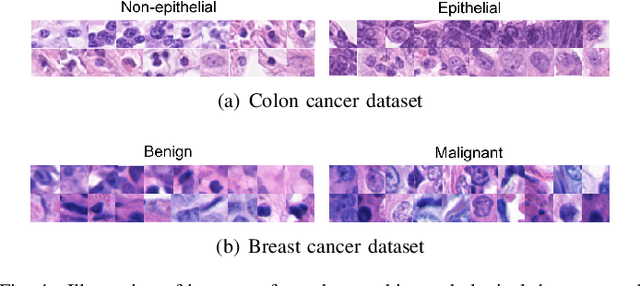
Weakly supervised instance labeling using only image-level labels, in lieu of expensive fine-grained pixel annotations, is crucial in several applications including medical image analysis. In contrast to conventional instance segmentation scenarios in computer vision, the problems that we consider are characterized by a small number of training images and non-local patterns that lead to the diagnosis. In this paper, we explore the use of multiple instance learning (MIL) to design an instance label generator under this weakly supervised setting. Motivated by the observation that an MIL model can handle bags of varying sizes, we propose to repurpose an MIL model originally trained for bag-level classification to produce reliable predictions for single instances, i.e., bags of size $1$. To this end, we introduce a novel regularization strategy based on virtual adversarial training for improving MIL training, and subsequently develop a knowledge distillation technique for repurposing the trained MIL model. Using empirical studies on colon cancer and breast cancer detection from histopathological images, we show that the proposed approach produces high-quality instance-level prediction and significantly outperforms state-of-the MIL methods.
Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging
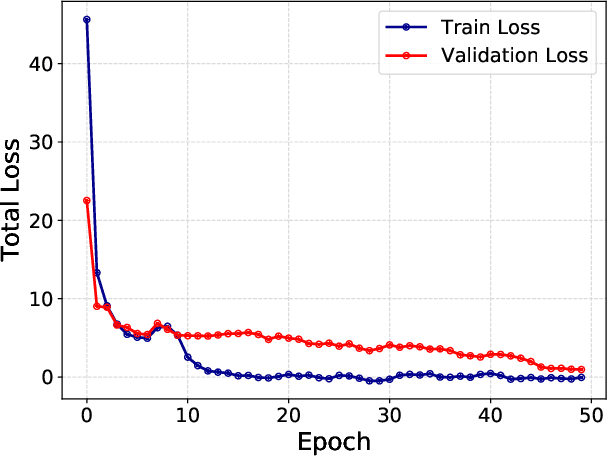
Sep 27, 2019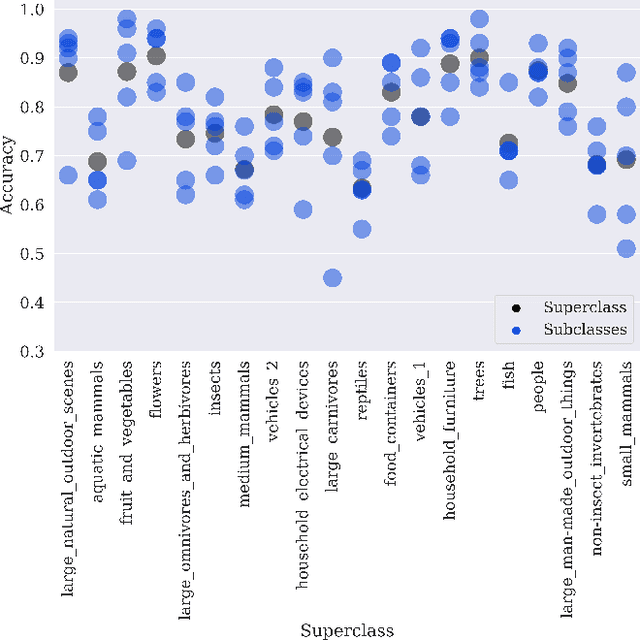
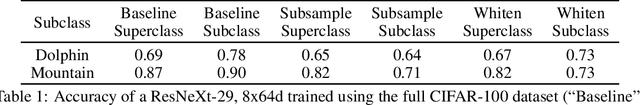
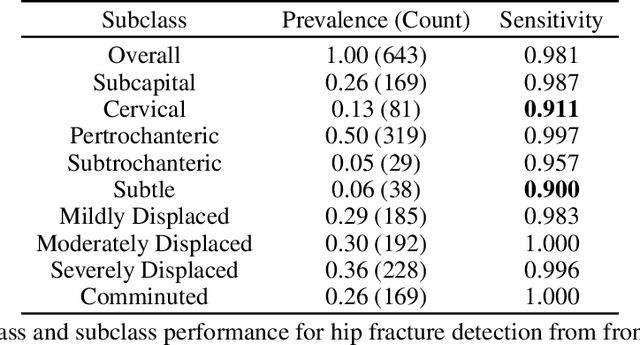
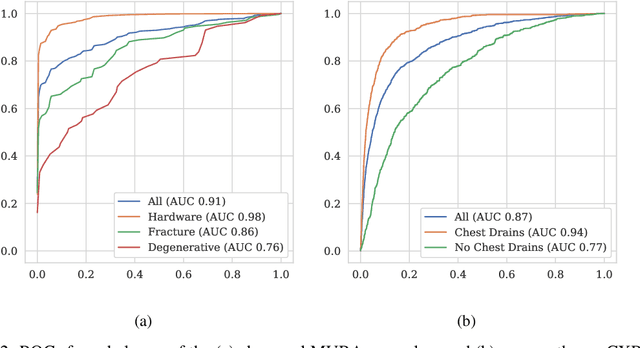
Machine learning models for medical image analysis often suffer from poor performance on important subsets of a population that are not identified during training or testing. For example, overall performance of a cancer detection model may be high, but the model still consistently misses a rare but aggressive cancer subtype. We refer to this problem as hidden stratification, and observe that it results from incompletely describing the meaningful variation in a dataset. While hidden stratification can substantially reduce the clinical efficacy of machine learning models, its effects remain difficult to measure. In this work, we assess the utility of several possible techniques for measuring and describing hidden stratification effects, and characterize these effects both on multiple medical imaging datasets and via synthetic experiments on the well-characterised CIFAR-100 benchmark dataset. We find evidence that hidden stratification can occur in unidentified imaging subsets with low prevalence, low label quality, subtle distinguishing features, or spurious correlates, and that it can result in relative performance differences of over 20% on clinically important subsets. Finally, we explore the clinical implications of our findings, and suggest that evaluation of hidden stratification should be a critical component of any machine learning deployment in medical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge