"cancer detection": models, code, and papers
MHSnet: Multi-head and Spatial Attention Network with False-Positive Reduction for Pulmonary Nodules Detection
Feb 05, 2022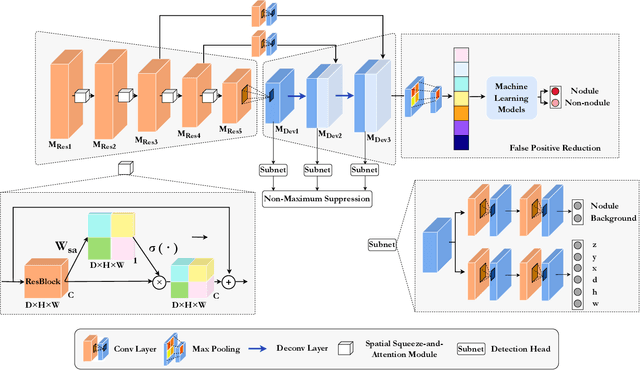
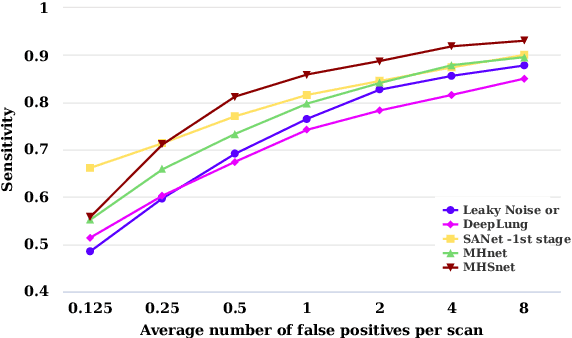
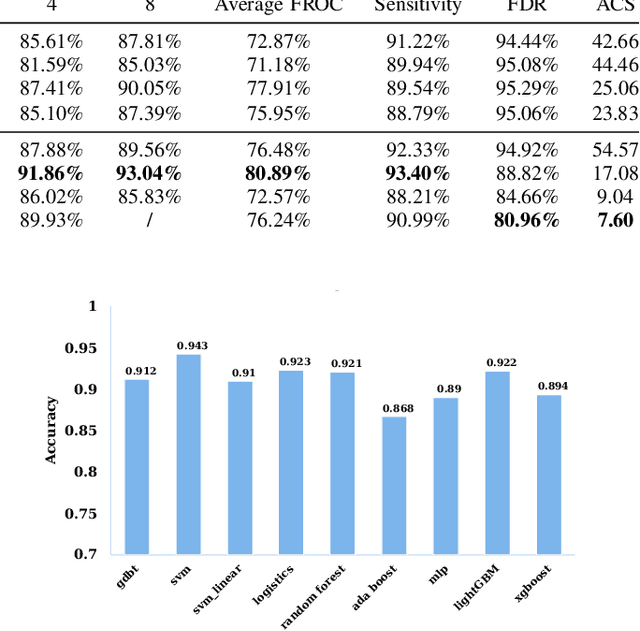
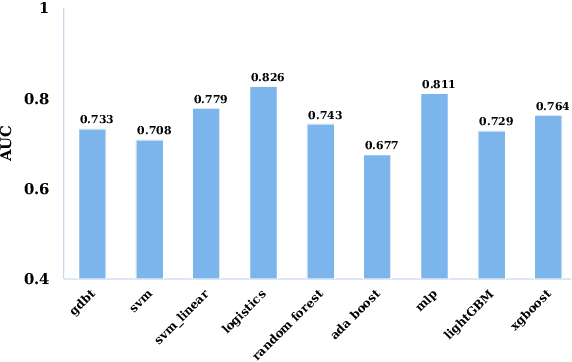
The mortality of lung cancer has ranked high among cancers for many years. Early detection of lung cancer is critical for disease prevention, cure, and mortality rate reduction. However, existing detection methods on pulmonary nodules introduce an excessive number of false positive proposals in order to achieve high sensitivity, which is not practical in clinical situations. In this paper, we propose the multi-head detection and spatial squeeze-and-attention network, MHSnet, to detect pulmonary nodules, in order to aid doctors in the early diagnosis of lung cancers. Specifically, we first introduce multi-head detectors and skip connections to customize for the variety of nodules in sizes, shapes and types and capture multi-scale features. Then, we implement a spatial attention module to enable the network to focus on different regions differently inspired by how experienced clinicians screen CT images, which results in fewer false positive proposals. Lastly, we present a lightweight but effective false positive reduction module with the Linear Regression model to cut down the number of false positive proposals, without any constraints on the front network. Extensive experimental results compared with the state-of-the-art models have shown the superiority of the MHSnet in terms of the average FROC, sensitivity and especially false discovery rate (2.98% and 2.18% improvement in terms of average FROC and sensitivity, 5.62% and 28.33% decrease in terms of false discovery rate and average candidates per scan). The false positive reduction module significantly decreases the average number of candidates generated per scan by 68.11% and the false discovery rate by 13.48%, which is promising to reduce distracted proposals for the downstream tasks based on the detection results.
Self-Supervised U-Net for Segmenting Flat and Sessile Polyps
Oct 17, 2021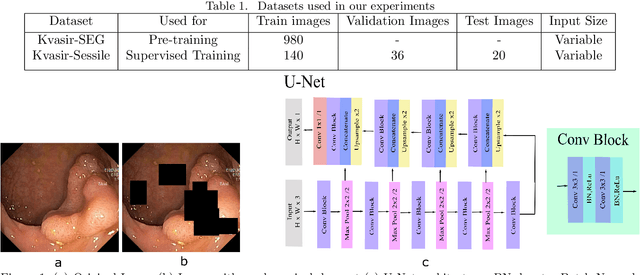
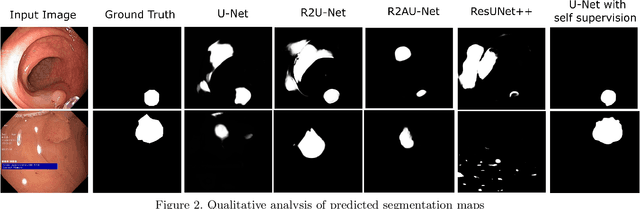
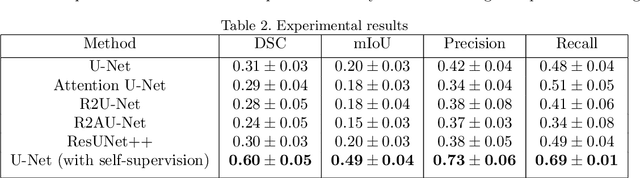
Colorectal Cancer(CRC) poses a great risk to public health. It is the third most common cause of cancer in the US. Development of colorectal polyps is one of the earliest signs of cancer. Early detection and resection of polyps can greatly increase survival rate to 90%. Manual inspection can cause misdetections because polyps vary in color, shape, size and appearance. To this end, Computer-Aided Diagnosis systems(CADx) has been proposed that detect polyps by processing the colonoscopic videos. The system acts a secondary check to help clinicians reduce misdetections so that polyps may be resected before they transform to cancer. Polyps vary in color, shape, size, texture and appearance. As a result, the miss rate of polyps is between 6% and 27% despite the prominence of CADx solutions. Furthermore, sessile and flat polyps which have diameter less than 10 mm are more likely to be undetected. Convolutional Neural Networks(CNN) have shown promising results in polyp segmentation. However, all of these works have a supervised approach and are limited by the size of the dataset. It was observed that smaller datasets reduce the segmentation accuracy of ResUNet++. We train a U-Net to inpaint randomly dropped out pixels in the image as a proxy task. The dataset we use for pre-training is Kvasir-SEG dataset. This is followed by a supervised training on the limited Kvasir-Sessile dataset. Our experimental results demonstrate that with limited annotated dataset and a larger unlabeled dataset, self-supervised approach is a better alternative than fully supervised approach. Specifically, our self-supervised U-Net performs better than five segmentation models which were trained in supervised manner on the Kvasir-Sessile dataset.
Colon Nuclei Instance Segmentation using a Probabilistic Two-Stage Detector
Mar 01, 2022
Cancer is one of the leading causes of death in the developed world. Cancer diagnosis is performed through the microscopic analysis of a sample of suspicious tissue. This process is time consuming and error prone, but Deep Learning models could be helpful for pathologists during cancer diagnosis. We propose to change the CenterNet2 object detection model to also perform instance segmentation, which we call SegCenterNet2. We train SegCenterNet2 in the CoNIC challenge dataset and show that it performs better than Mask R-CNN in the competition metrics.
Computer-aided Tumor Diagnosis in Automated Breast Ultrasound using 3D Detection Network
Jul 31, 2020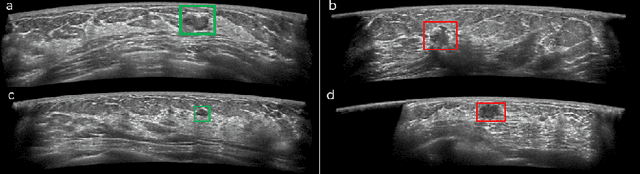
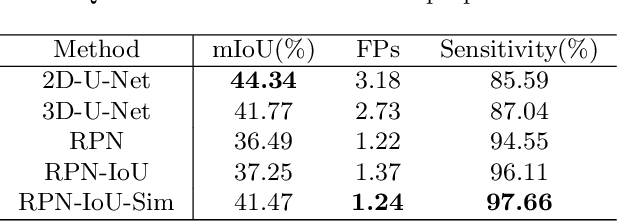
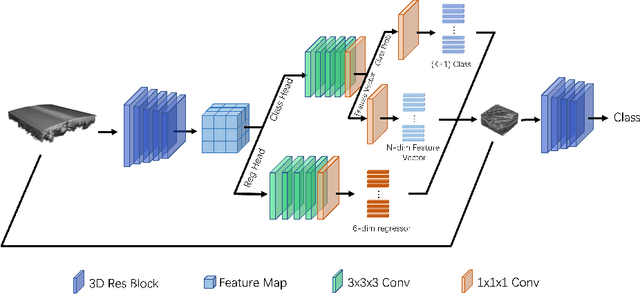
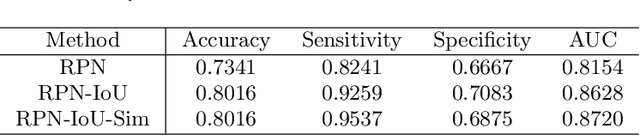
Automated breast ultrasound (ABUS) is a new and promising imaging modality for breast cancer detection and diagnosis, which could provide intuitive 3D information and coronal plane information with great diagnostic value. However, manually screening and diagnosing tumors from ABUS images is very time-consuming and overlooks of abnormalities may happen. In this study, we propose a novel two-stage 3D detection network for locating suspected lesion areas and further classifying lesions as benign or malignant tumors. Specifically, we propose a 3D detection network rather than frequently-used segmentation network to locate lesions in ABUS images, thus our network can make full use of the spatial context information in ABUS images. A novel similarity loss is designed to effectively distinguish lesions from background. Then a classification network is employed to identify the located lesions as benign or malignant. An IoU-balanced classification loss is adopted to improve the correlation between classification and localization task. The efficacy of our network is verified from a collected dataset of 418 patients with 145 benign tumors and 273 malignant tumors. Experiments show our network attains a sensitivity of 97.66% with 1.23 false positives (FPs), and has an area under the curve(AUC) value of 0.8720.
Unsupervised Contrastive Learning based Transformer for Lung Nodule Detection
Apr 30, 2022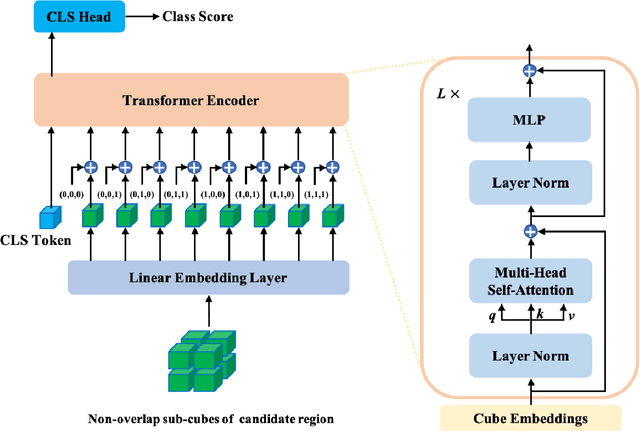
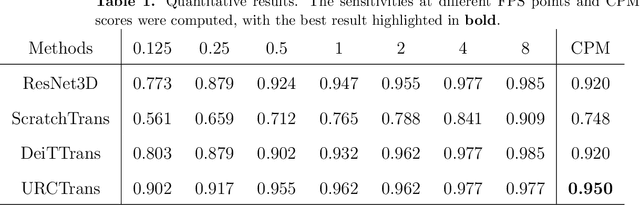
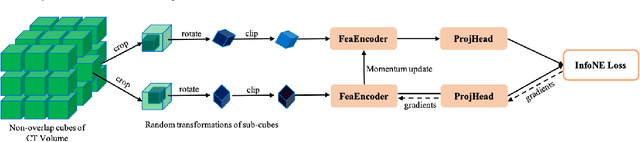
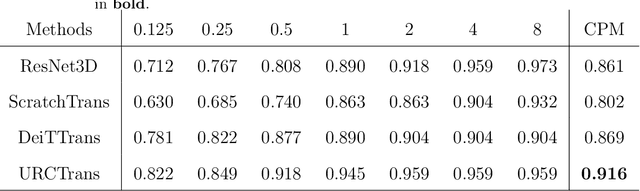
Early detection of lung nodules with computed tomography (CT) is critical for the longer survival of lung cancer patients and better quality of life. Computer-aided detection/diagnosis (CAD) is proven valuable as a second or concurrent reader in this context. However, accurate detection of lung nodules remains a challenge for such CAD systems and even radiologists due to not only the variability in size, location, and appearance of lung nodules but also the complexity of lung structures. This leads to a high false-positive rate with CAD, compromising its clinical efficacy. Motivated by recent computer vision techniques, here we present a self-supervised region-based 3D transformer model to identify lung nodules among a set of candidate regions. Specifically, a 3D vision transformer (ViT) is developed that divides a CT image volume into a sequence of non-overlap cubes, extracts embedding features from each cube with an embedding layer, and analyzes all embedding features with a self-attention mechanism for the prediction. To effectively train the transformer model on a relatively small dataset, the region-based contrastive learning method is used to boost the performance by pre-training the 3D transformer with public CT images. Our experiments show that the proposed method can significantly improve the performance of lung nodule screening in comparison with the commonly used 3D convolutional neural networks.
Self-Rule to Adapt: Generalized Multi-source Feature Learning Using Unsupervised Domain Adaptation for Colorectal Cancer Tissue Detection
Aug 20, 2021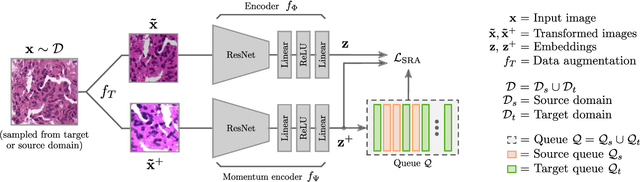
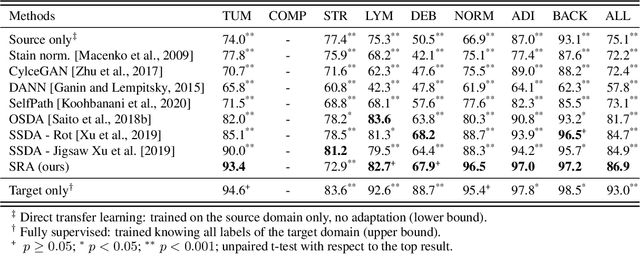
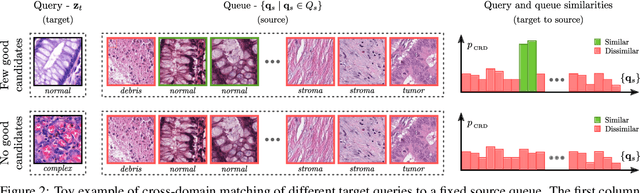
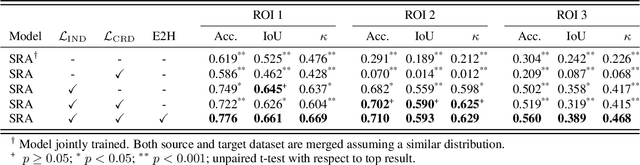
Supervised learning is constrained by the availability of labeled data, which are especially expensive to acquire in the field of digital pathology. Making use of open-source data for pre-training or using domain adaptation can be a way to overcome this issue. However, pre-trained networks often fail to generalize to new test domains that are not distributed identically due to variations in tissue stainings, types, and textures. Additionally, current domain adaptation methods mainly rely on fully-labeled source datasets. In this work, we propose SRA, which takes advantage of self-supervised learning to perform domain adaptation and removes the necessity of a fully-labeled source dataset. SRA can effectively transfer the discriminative knowledge obtained from a few labeled source domain's data to a new target domain without requiring additional tissue annotations. Our method harnesses both domains' structures by capturing visual similarity with intra-domain and cross-domain self-supervision. Moreover, we present a generalized formulation of our approach that allows the architecture to learn from multi-source domains. We show that our proposed method outperforms baselines for domain adaptation of colorectal tissue type classification and further validate our approach on our in-house clinical cohort. The code and models are available open-source: https://github.com/christianabbet/SRA.
Deep learning in magnetic resonance prostate segmentation: A review and a new perspective
Nov 16, 2020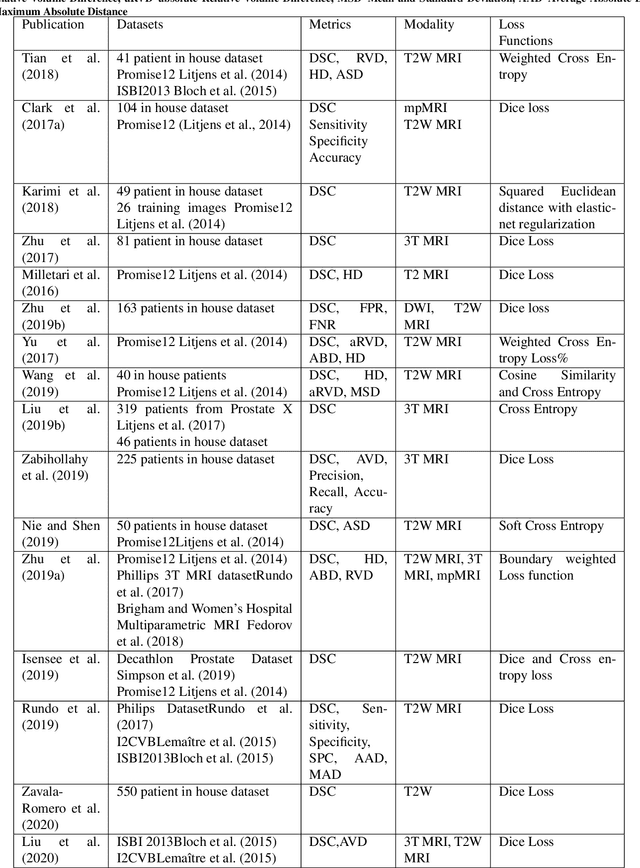
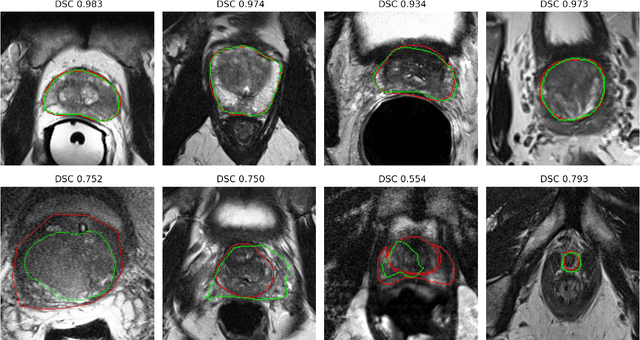
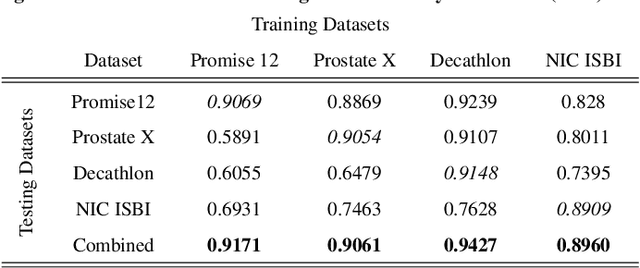
Prostate radiotherapy is a well established curative oncology modality, which in future will use Magnetic Resonance Imaging (MRI)-based radiotherapy for daily adaptive radiotherapy target definition. However the time needed to delineate the prostate from MRI data accurately is a time consuming process. Deep learning has been identified as a potential new technology for the delivery of precision radiotherapy in prostate cancer, where accurate prostate segmentation helps in cancer detection and therapy. However, the trained models can be limited in their application to clinical setting due to different acquisition protocols, limited publicly available datasets, where the size of the datasets are relatively small. Therefore, to explore the field of prostate segmentation and to discover a generalisable solution, we review the state-of-the-art deep learning algorithms in MR prostate segmentation; provide insights to the field by discussing their limitations and strengths; and propose an optimised 2D U-Net for MR prostate segmentation. We evaluate the performance on four publicly available datasets using Dice Similarity Coefficient (DSC) as performance metric. Our experiments include within dataset evaluation and cross-dataset evaluation. The best result is achieved by composite evaluation (DSC of 0.9427 on Decathlon test set) and the poorest result is achieved by cross-dataset evaluation (DSC of 0.5892, Prostate X training set, Promise 12 testing set). We outline the challenges and provide recommendations for future work. Our research provides a new perspective to MR prostate segmentation and more importantly, we provide standardised experiment settings for researchers to evaluate their algorithms. Our code is available at https://github.com/AIEMMU/MRI\_Prostate.
Explainable Disease Classification via weakly-supervised segmentation
Aug 24, 2020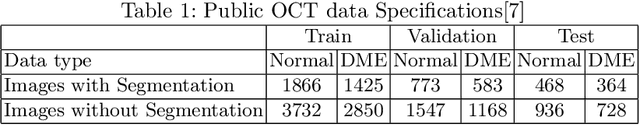
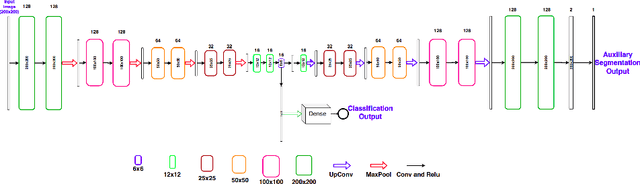
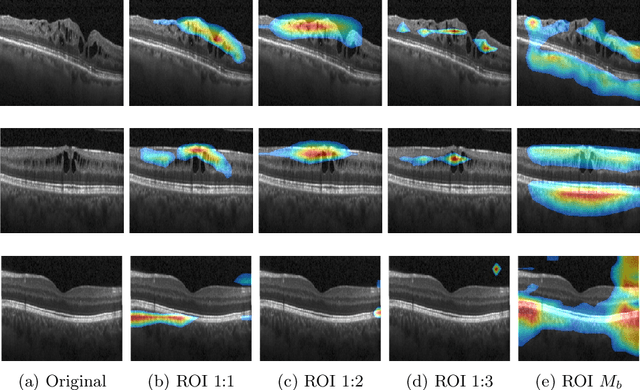
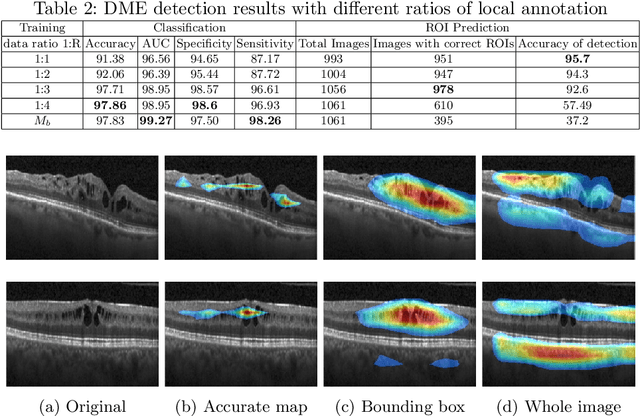
Deep learning based approaches to Computer Aided Diagnosis (CAD) typically pose the problem as an image classification (Normal or Abnormal) problem. These systems achieve high to very high accuracy in specific disease detection for which they are trained but lack in terms of an explanation for the provided decision/classification result. The activation maps which correspond to decisions do not correlate well with regions of interest for specific diseases. This paper examines this problem and proposes an approach which mimics the clinical practice of looking for an evidence prior to diagnosis. A CAD model is learnt using a mixed set of information: class labels for the entire training set of images plus a rough localisation of suspect regions as an extra input for a smaller subset of training images for guiding the learning. The proposed approach is illustrated with detection of diabetic macular edema (DME) from OCT slices. Results of testing on on a large public dataset show that with just a third of images with roughly segmented fluid filled regions, the classification accuracy is on par with state of the art methods while providing a good explanation in the form of anatomically accurate heatmap /region of interest. The proposed solution is then adapted to Breast Cancer detection from mammographic images. Good evaluation results on public datasets underscores the generalisability of the proposed solution.
Skin cancer detection based on deep learning and entropy to detect outlier samples
Sep 10, 2019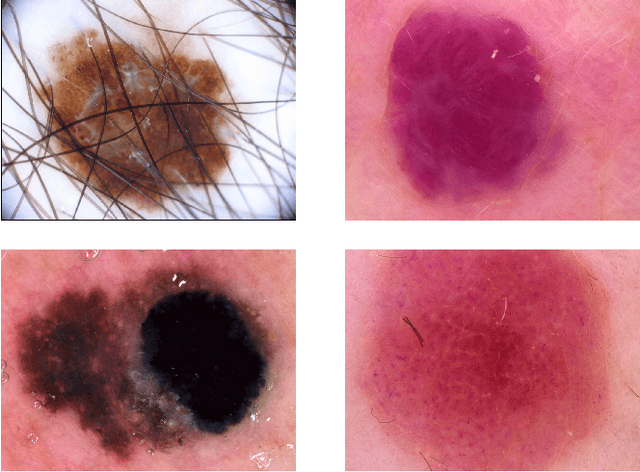
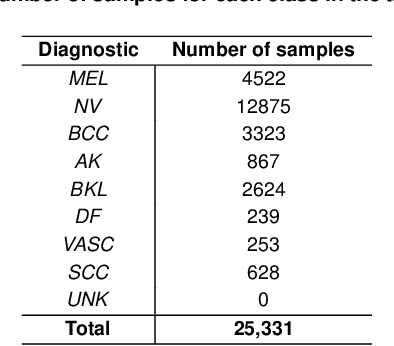
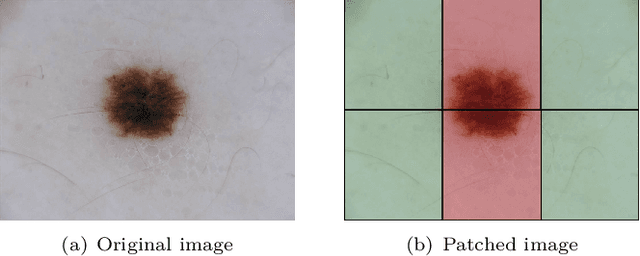
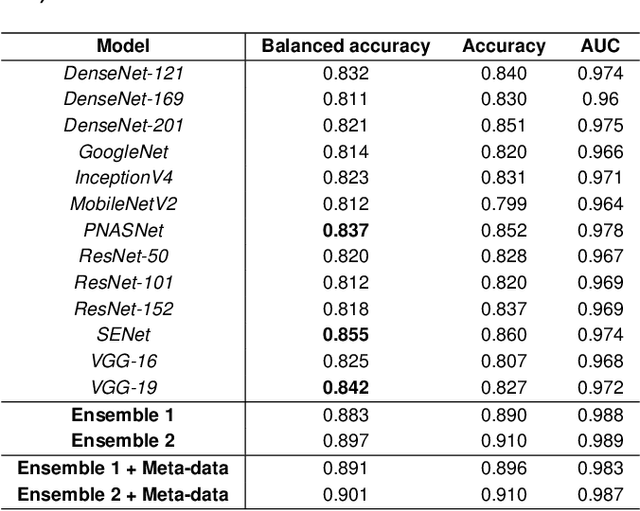
We describe our methods to address both tasks of the ISIC 2019 challenge. The goal of this challenge is to provide the diagnostic for skin cancer using images and meta-data. There are nine classes in the dataset, nonetheless, one of them is an outlier and is not present on it. To tackle the challenge, we apply an ensemble of classifiers, which has 13 convolutional neural networks (CNN), we develop two approaches to handle the outlier class and we propose a straightforward method to use the meta-data along with the images. Throughout this report, we detail each methodology and parameters to make it easy to replicate our work. The results obtained are in accordance with the previous challenges and the approaches to detect the outlier class and to address the meta-data seem to be work properly.
Detection, growth quantification and malignancy prediction of pulmonary nodules using deep convolutional networks in follow-up CT scans
Mar 26, 2021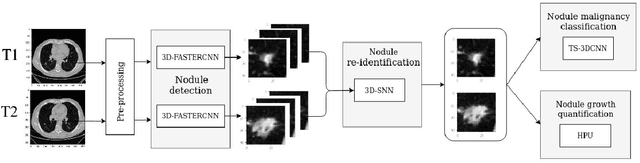
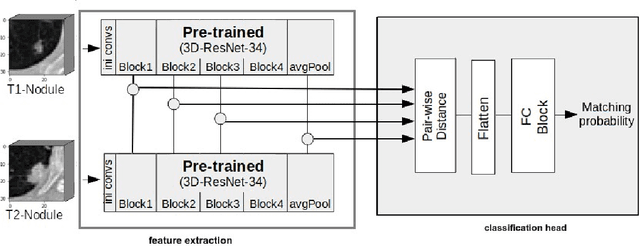
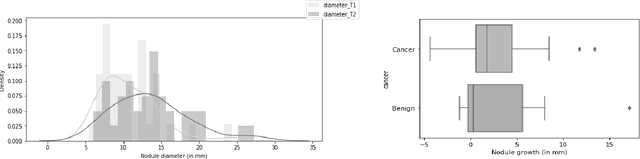
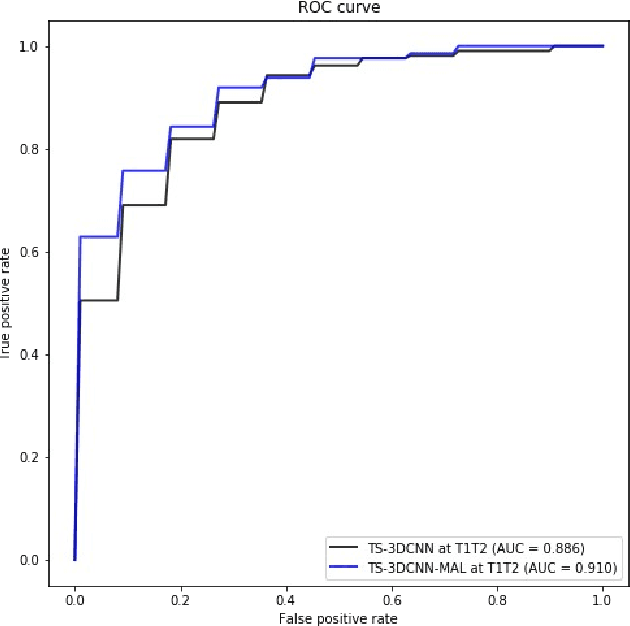
We address the problem of supporting radiologists in the longitudinal management of lung cancer. Therefore, we proposed a deep learning pipeline, composed of four stages that completely automatized from the detection of nodules to the classification of cancer, through the detection of growth in the nodules. In addition, the pipeline integrated a novel approach for nodule growth detection, which relied on a recent hierarchical probabilistic U-Net adapted to report uncertainty estimates. Also, a second novel method was introduced for lung cancer nodule classification, integrating into a two stream 3D-CNN network the estimated nodule malignancy probabilities derived from a pretrained nodule malignancy network. The pipeline was evaluated in a longitudinal cohort and reported comparable performances to the state of art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge