"cancer detection": models, code, and papers
End-to-end Prostate Cancer Detection in bpMRI via 3D CNNs: Effect of Attention Mechanisms, Clinical Priori and Decoupled False Positive Reduction
Jan 22, 2021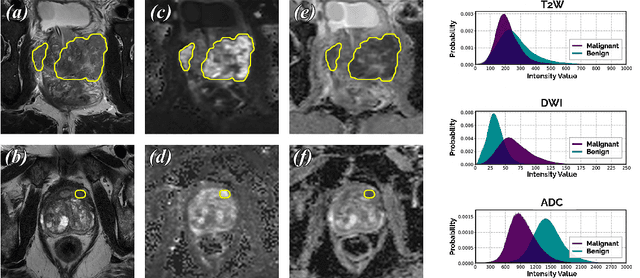
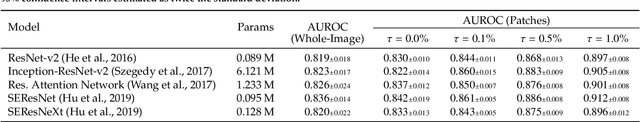
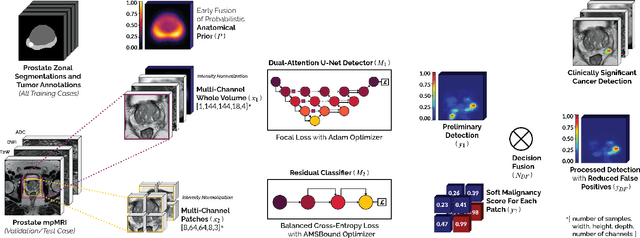
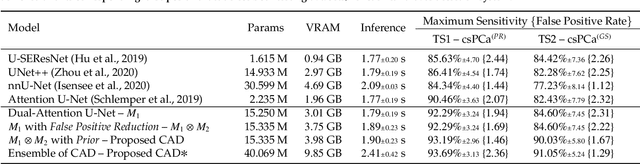
We present a novel multi-stage 3D computer-aided detection and diagnosis (CAD) model for automated localization of clinically significant prostate cancer (csPCa) in bi-parametric MR imaging (bpMRI). State-of-the-art attention mechanisms drive its detection network, which aims to accurately discriminate csPCa lesions from indolent cancer and the wide range of benign pathology that can afflict the prostate gland. In parallel, a decoupled residual classifier is used to achieve consistent false positive reduction, without sacrificing high detection sensitivity or computational efficiency. Furthermore, a probabilistic anatomical prior, which captures the spatial prevalence of csPCa and its zonal distinction, is computed and encoded into the CNN architecture to guide model generalization with domain-specific clinical knowledge. For 486 institutional testing scans, the 3D CAD system achieves $83.69\pm5.22\%$ and $93.19\pm2.96\%$ detection sensitivity at 0.50 and 1.46 false positive(s) per patient, respectively, along with $0.882$ AUROC in patient-based diagnosis $-$significantly outperforming four state-of-the-art baseline architectures (USEResNet, UNet++, nnU-Net, Attention U-Net) from recent literature. For 296 external testing scans, the ensembled CAD system shares moderate agreement with a consensus of expert radiologists ($76.69\%$; $kappa=0.511$) and independent pathologists ($81.08\%$; $kappa=0.559$); demonstrating a strong ability to localize histologically-confirmed malignancies and generalize beyond the radiologically-estimated annotations of the 1950 training-validation cases used in this study.
MHSnet: Multi-head and Spatial Attention Network with False-Positive Reduction for Pulmonary Nodules Detection
Feb 16, 2022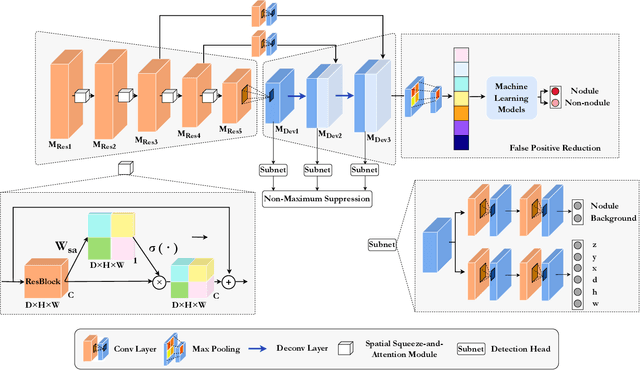
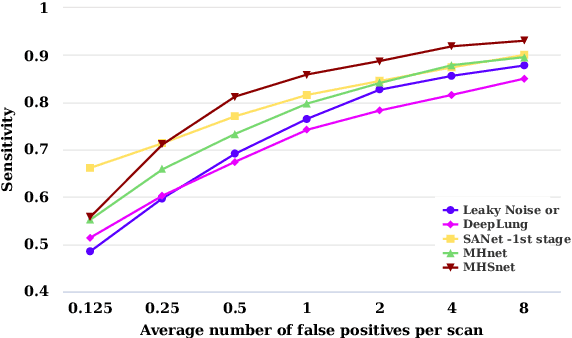
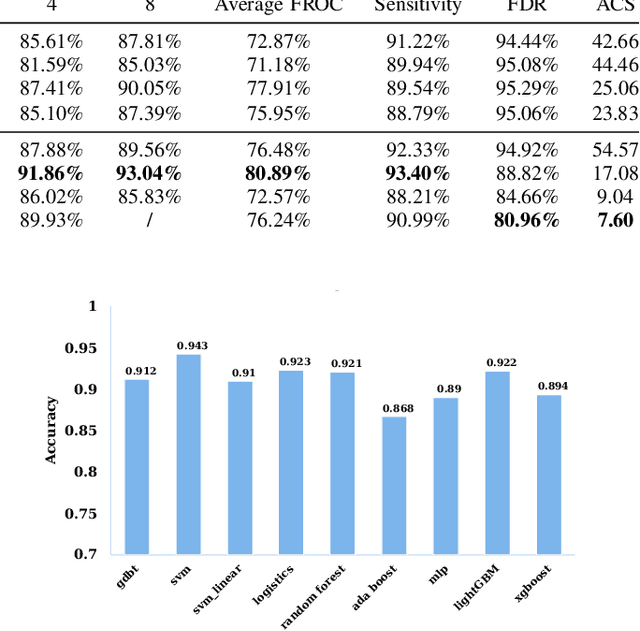
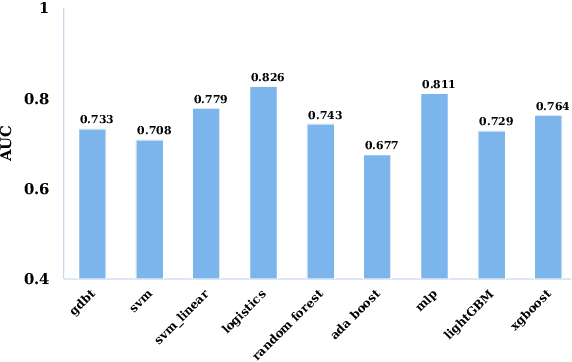
The mortality of lung cancer has ranked high among cancers for many years. Early detection of lung cancer is critical for disease prevention, cure, and mortality rate reduction. However, existing detection methods on pulmonary nodules introduce an excessive number of false positive proposals in order to achieve high sensitivity, which is not practical in clinical situations. In this paper, we propose the multi-head detection and spatial squeeze-and-attention network, MHSnet, to detect pulmonary nodules, in order to aid doctors in the early diagnosis of lung cancers. Specifically, we first introduce multi-head detectors and skip connections to customize for the variety of nodules in sizes, shapes and types and capture multi-scale features. Then, we implement a spatial attention module to enable the network to focus on different regions differently inspired by how experienced clinicians screen CT images, which results in fewer false positive proposals. Lastly, we present a lightweight but effective false positive reduction module with the Linear Regression model to cut down the number of false positive proposals, without any constraints on the front network. Extensive experimental results compared with the state-of-the-art models have shown the superiority of the MHSnet in terms of the average FROC, sensitivity and especially false discovery rate (2.98% and 2.18% improvement in terms of average FROC and sensitivity, 5.62% and 28.33% decrease in terms of false discovery rate and average candidates per scan). The false positive reduction module significantly decreases the average number of candidates generated per scan by 68.11% and the false discovery rate by 13.48%, which is promising to reduce distracted proposals for the downstream tasks based on the detection results.
Overcoming the limitations of patch-based learning to detect cancer in whole slide images
Dec 01, 2020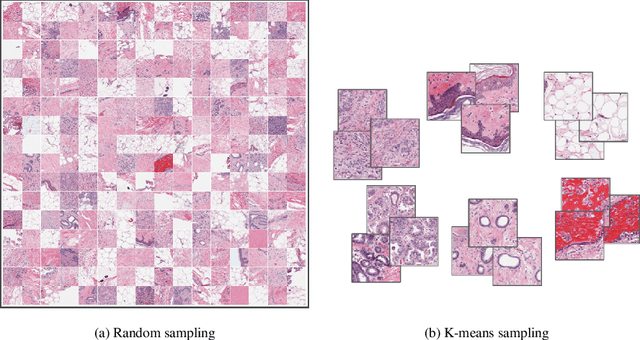
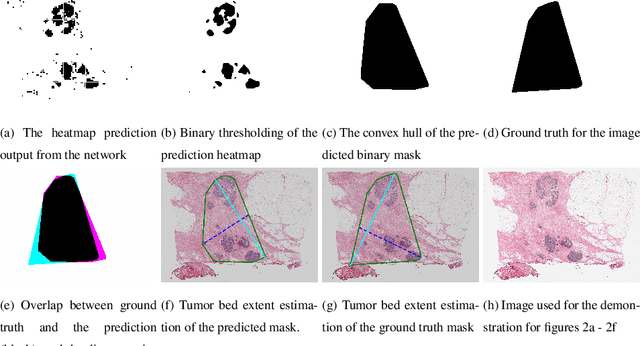
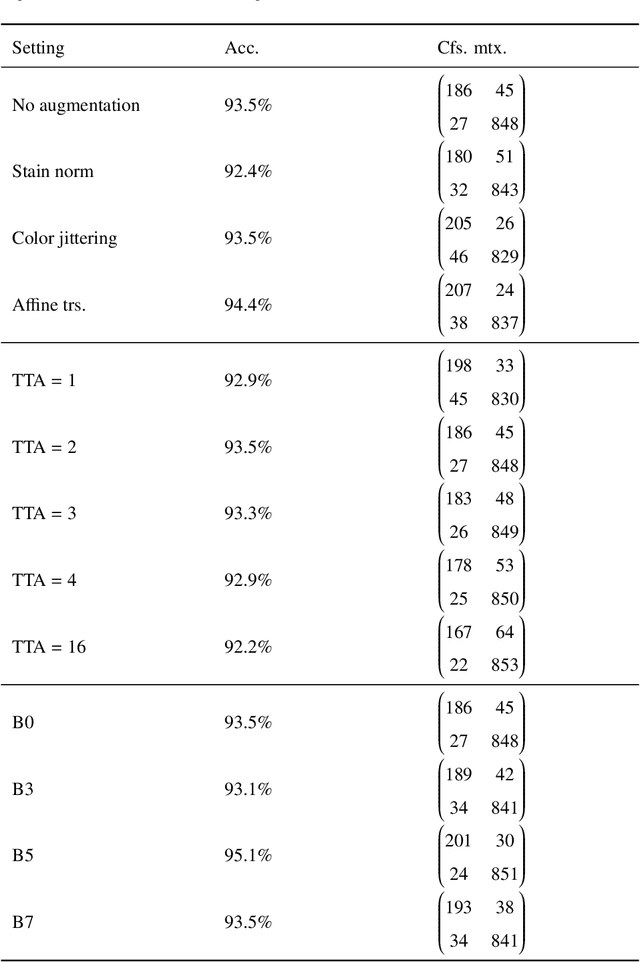
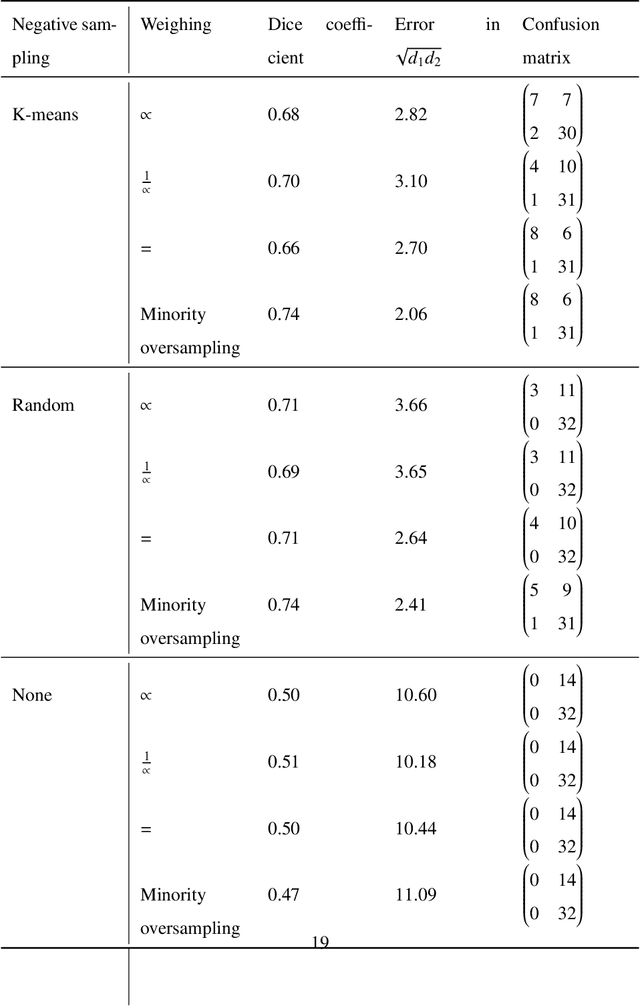
Whole slide images (WSIs) pose unique challenges when training deep learning models. They are very large which makes it necessary to break each image down into smaller patches for analysis, image features have to be extracted at multiple scales in order to capture both detail and context, and extreme class imbalances may exist. Significant progress has been made in the analysis of these images, thanks largely due to the availability of public annotated datasets. We postulate, however, that even if a method scores well on a challenge task, this success may not translate to good performance in a more clinically relevant workflow. Many datasets consist of image patches which may suffer from data curation bias; other datasets are only labelled at the whole slide level and the lack of annotations across an image may mask erroneous local predictions so long as the final decision is correct. In this paper, we outline the differences between patch or slide-level classification versus methods that need to localize or segment cancer accurately across the whole slide, and we experimentally verify that best practices differ in both cases. We apply a binary cancer detection network on post neoadjuvant therapy breast cancer WSIs to find the tumor bed outlining the extent of cancer, a task which requires sensitivity and precision across the whole slide. We extensively study multiple design choices and their effects on the outcome, including architectures and augmentations. Furthermore, we propose a negative data sampling strategy, which drastically reduces the false positive rate (7% on slide level) and improves each metric pertinent to our problem, with a 15% reduction in the error of tumor extent.
2D Convolutional Neural Networks for 3D Digital Breast Tomosynthesis Classification
Feb 27, 2020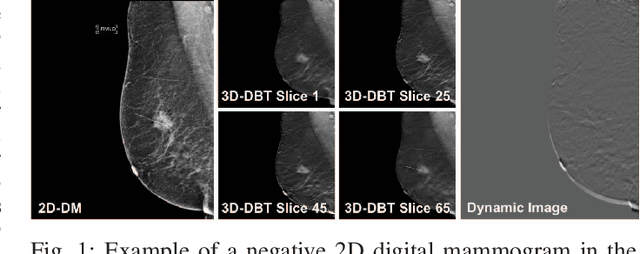
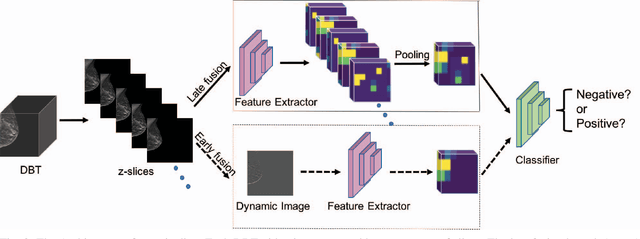
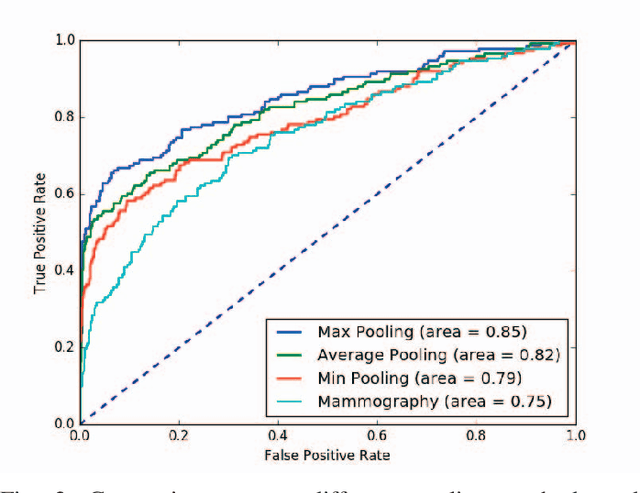
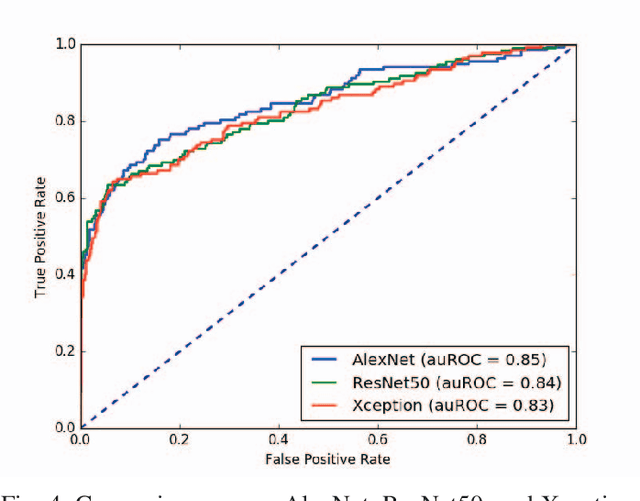
Automated methods for breast cancer detection have focused on 2D mammography and have largely ignored 3D digital breast tomosynthesis (DBT), which is frequently used in clinical practice. The two key challenges in developing automated methods for DBT classification are handling the variable number of slices and retaining slice-to-slice changes. We propose a novel deep 2D convolutional neural network (CNN) architecture for DBT classification that simultaneously overcomes both challenges. Our approach operates on the full volume, regardless of the number of slices, and allows the use of pre-trained 2D CNNs for feature extraction, which is important given the limited amount of annotated training data. In an extensive evaluation on a real-world clinical dataset, our approach achieves 0.854 auROC, which is 28.80% higher than approaches based on 3D CNNs. We also find that these improvements are stable across a range of model configurations.
Classification of Histopathology Images of Lung Cancer Using Convolutional Neural Network (CNN)
Dec 27, 2021
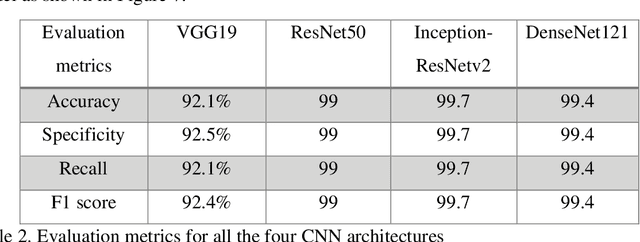
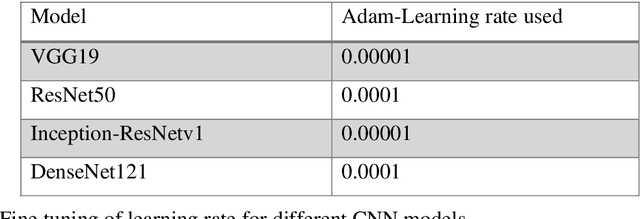
Cancer is the uncontrollable cell division of abnormal cells inside the human body, which can spread to other body organs. It is one of the non-communicable diseases (NCDs) and NCDs accounts for 71% of total deaths worldwide whereas lung cancer is the second most diagnosed cancer after female breast cancer. Cancer survival rate of lung cancer is only 19%. There are various methods for the diagnosis of lung cancer, such as X-ray, CT scan, PET-CT scan, bronchoscopy and biopsy. However, to know the subtype of lung cancer based on the tissue type H and E staining is widely used, where the staining is done on the tissue aspirated from a biopsy. Studies have reported that the type of histology is associated with prognosis and treatment in lung cancer. Therefore, early and accurate detection of lung cancer histology is an urgent need and as its treatment is dependent on the type of histology, molecular profile and stage of the disease, it is most essential to analyse the histopathology images of lung cancer. Hence, to speed up the vital process of diagnosis of lung cancer and reduce the burden on pathologists, Deep learning techniques are used. These techniques have shown improved efficacy in the analysis of histopathology slides of cancer. Several studies reported the importance of convolution neural networks (CNN) in the classification of histopathological pictures of various cancer types such as brain, skin, breast, lung, colorectal cancer. In this study tri-category classification of lung cancer images (normal, adenocarcinoma and squamous cell carcinoma) are carried out by using ResNet 50, VGG-19, Inception_ResNet_V2 and DenseNet for the feature extraction and triplet loss to guide the CNN such that it increases inter-cluster distance and reduces intra-cluster distance.
Colon Nuclei Instance Segmentation using a Probabilistic Two-Stage Detector
Mar 01, 2022
Cancer is one of the leading causes of death in the developed world. Cancer diagnosis is performed through the microscopic analysis of a sample of suspicious tissue. This process is time consuming and error prone, but Deep Learning models could be helpful for pathologists during cancer diagnosis. We propose to change the CenterNet2 object detection model to also perform instance segmentation, which we call SegCenterNet2. We train SegCenterNet2 in the CoNIC challenge dataset and show that it performs better than Mask R-CNN in the competition metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge