"cancer detection": models, code, and papers
Machine Learning Applications in Lung Cancer Diagnosis, Treatment and Prognosis
Mar 26, 2022
The recent development of imaging and sequencing technologies enables systematic advances in the clinical study of lung cancer. Meanwhile, the human mind is limited in effectively handling and fully utilizing the accumulation of such enormous amounts of data. Machine learning-based approaches play a critical role in integrating and analyzing these large and complex datasets, which have extensively characterized lung cancer through the use of different perspectives from these accrued data. In this article, we provide an overview of machine learning-based approaches that strengthen the varying aspects of lung cancer diagnosis and therapy, including early detection, auxiliary diagnosis, prognosis prediction and immunotherapy practice. Moreover, we highlight the challenges and opportunities for future applications of machine learning in lung cancer.
The impact of using voxel-level segmentation metrics on evaluating multifocal prostate cancer localisation
Mar 31, 2022
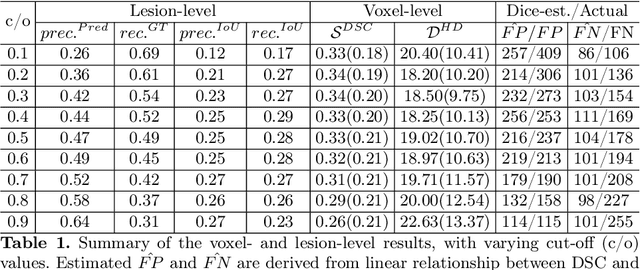
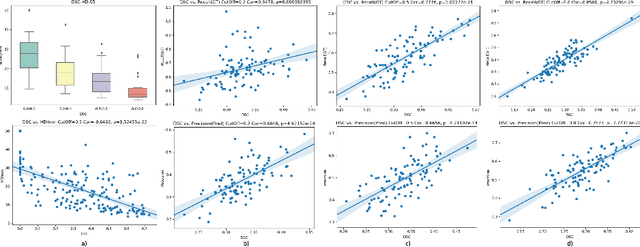
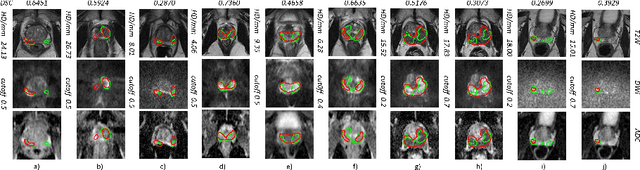
Dice similarity coefficient (DSC) and Hausdorff distance (HD) are widely used for evaluating medical image segmentation. They have also been criticised, when reported alone, for their unclear or even misleading clinical interpretation. DSCs may also differ substantially from HDs, due to boundary smoothness or multiple regions of interest (ROIs) within a subject. More importantly, either metric can also have a nonlinear, non-monotonic relationship with outcomes based on Type 1 and 2 errors, designed for specific clinical decisions that use the resulting segmentation. Whilst cases causing disagreement between these metrics are not difficult to postulate. This work first proposes a new asymmetric detection metric, adapting those used in object detection, for planning prostate cancer procedures. The lesion-level metrics is then compared with the voxel-level DSC and HD, whereas a 3D UNet is used for segmenting lesions from multiparametric MR (mpMR) images. Based on experimental results we report pairwise agreement and correlation 1) between DSC and HD, and 2) between voxel-level DSC and recall-controlled precision at lesion-level, with Cohen's [0.49, 0.61] and Pearson's [0.66, 0.76] (p-values}<0.001) at varying cut-offs. However, the differences in false-positives and false-negatives, between the actual errors and the perceived counterparts if DSC is used, can be as high as 152 and 154, respectively, out of the 357 test set lesions. We therefore carefully conclude that, despite of the significant correlations, voxel-level metrics such as DSC can misrepresent lesion-level detection accuracy for evaluating localisation of multifocal prostate cancer and should be interpreted with caution.
CELNet: Evidence Localization for Pathology Images using Weakly Supervised Learning
Sep 16, 2019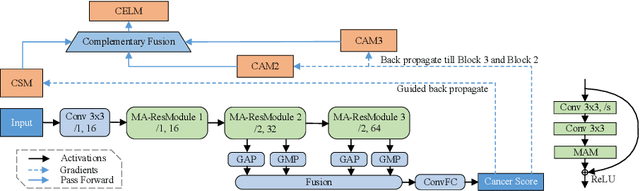
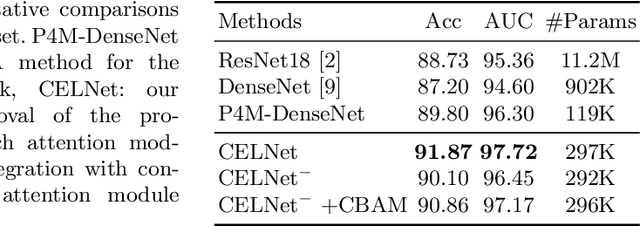
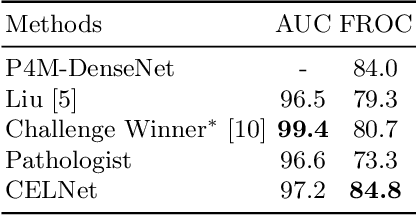
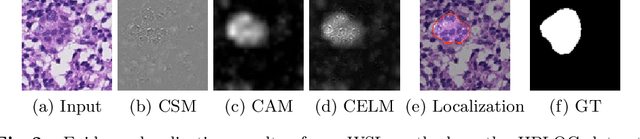
Despite deep convolutional neural networks boost the performance of image classification and segmentation in digital pathology analysis, they are usually weak in interpretability for clinical applications or require heavy annotations to achieve object localization. To overcome this problem, we propose a weakly supervised learning-based approach that can effectively learn to localize the discriminative evidence for a diagnostic label from weakly labeled training data. Experimental results show that our proposed method can reliably pinpoint the location of cancerous evidence supporting the decision of interest, while still achieving a competitive performance on glimpse-level and slide-level histopathologic cancer detection tasks.
Deep Learning-Based Automatic Detection of Poorly Positioned Mammograms to Minimize Patient Return Visits for Repeat Imaging: A Real-World Application
Sep 28, 2020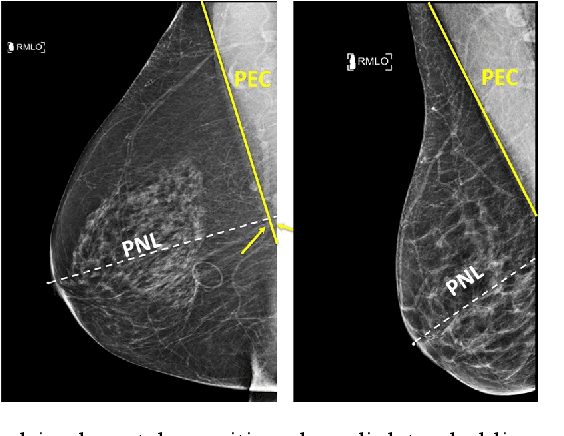
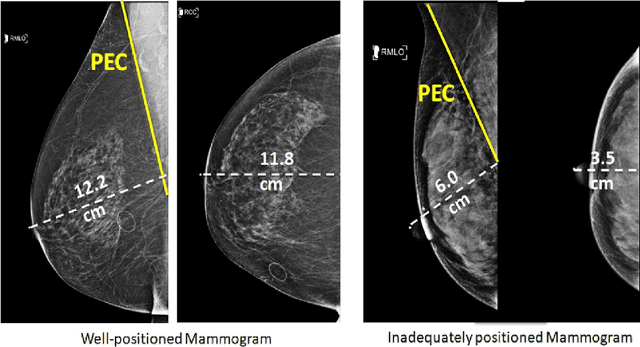
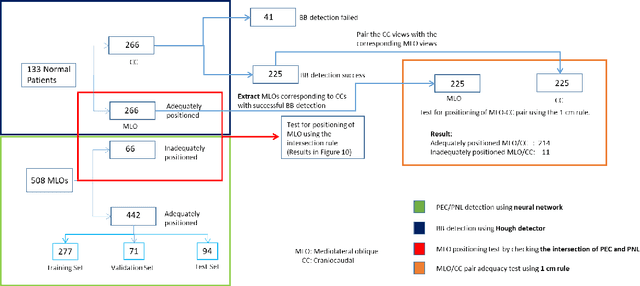

Screening mammograms are a routine imaging exam performed to detect breast cancer in its early stages to reduce morbidity and mortality attributed to this disease. In order to maximize the efficacy of breast cancer screening programs, proper mammographic positioning is paramount. Proper positioning ensures adequate visualization of breast tissue and is necessary for effective breast cancer detection. Therefore, breast-imaging radiologists must assess each mammogram for the adequacy of positioning before providing a final interpretation of the examination; this often necessitates return patient visits for additional imaging. In this paper, we propose a deep learning-algorithm method that mimics and automates this decision-making process to identify poorly positioned mammograms. Our objective for this algorithm is to assist mammography technologists in recognizing inadequately positioned mammograms real-time, improve the quality of mammographic positioning and performance, and ultimately reducing repeat visits for patients with initially inadequate imaging. The proposed model showed a true positive rate for detecting correct positioning of 91.35% in the mediolateral oblique view and 95.11% in the craniocaudal view. In addition to these results, we also present an automatically generated report which can aid the mammography technologist in taking corrective measures during the patient visit.
ESTAN: Enhanced Small Tumor-Aware Network for Breast Ultrasound Image Segmentation
Sep 27, 2020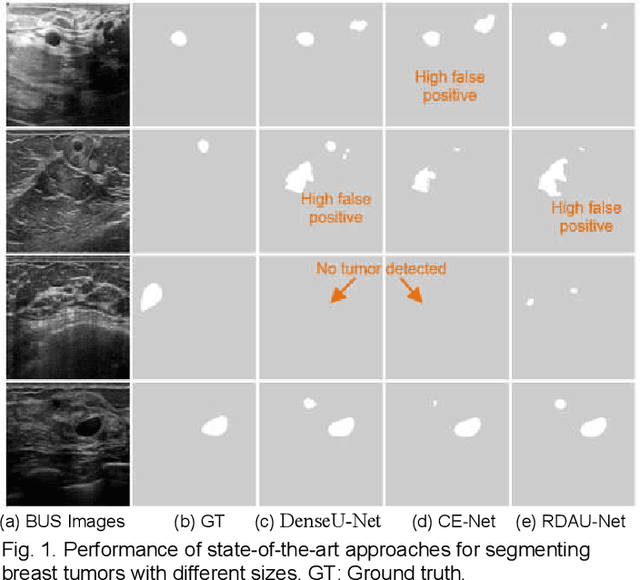
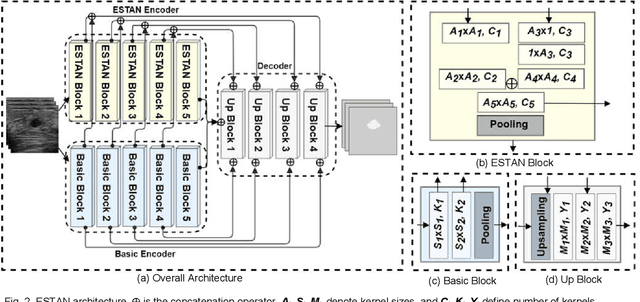
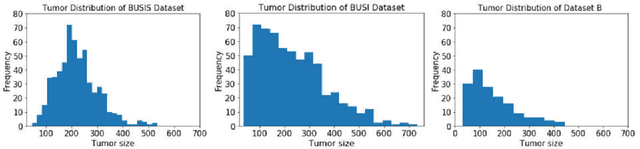
Breast tumor segmentation is a critical task in computer-aided diagnosis (CAD) systems for breast cancer detection because accurate tumor size, shape and location are important for further tumor quantification and classification. However, segmenting small tumors in ultrasound images is challenging, due to the speckle noise, varying tumor shapes and sizes among patients, and the existence of tumor-like image regions. Recently, deep learning-based approaches have achieved great success for biomedical image analysis, but current state-of-the-art approaches achieve poor performance for segmenting small breast tumors. In this paper, we propose a novel deep neural network architecture, namely Enhanced Small Tumor-Aware Network (ESTAN), to accurately and robustly segment breast tumors. ESTAN introduces two encoders to extract and fuse image context information at different scales and utilizes row-column-wise kernels in the encoder to adapt to breast anatomy. We validate the proposed approach and compare it to nine state-of-the-art approaches on three public breast ultrasound datasets using seven quantitative metrics. The results demonstrate that the proposed approach achieves the best overall performance and outperforms all other approaches on small tumor segmentation.
TransResU-Net: Transformer based ResU-Net for Real-Time Colonoscopy Polyp Segmentation
Jun 17, 2022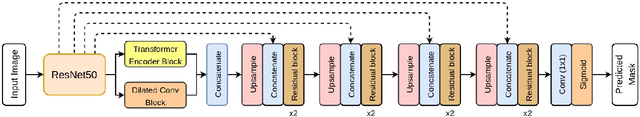
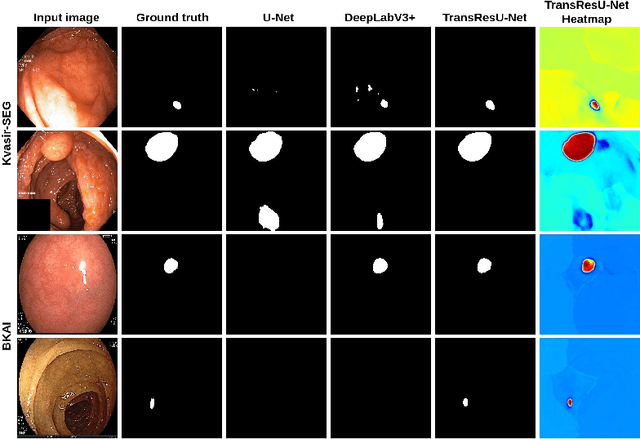
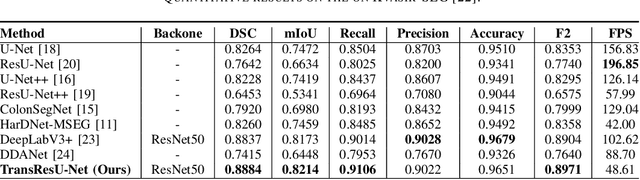
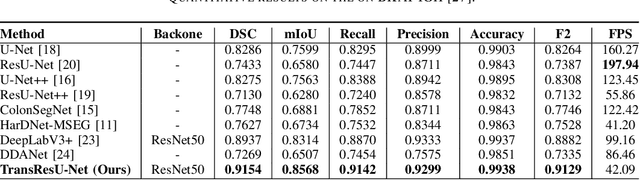
Colorectal cancer (CRC) is one of the most common causes of cancer and cancer-related mortality worldwide. Performing colon cancer screening in a timely fashion is the key to early detection. Colonoscopy is the primary modality used to diagnose colon cancer. However, the miss rate of polyps, adenomas and advanced adenomas remains significantly high. Early detection of polyps at the precancerous stage can help reduce the mortality rate and the economic burden associated with colorectal cancer. Deep learning-based computer-aided diagnosis (CADx) system may help gastroenterologists to identify polyps that may otherwise be missed, thereby improving the polyp detection rate. Additionally, CADx system could prove to be a cost-effective system that improves long-term colorectal cancer prevention. In this study, we proposed a deep learning-based architecture for automatic polyp segmentation, called Transformer ResU-Net (TransResU-Net). Our proposed architecture is built upon residual blocks with ResNet-50 as the backbone and takes the advantage of transformer self-attention mechanism as well as dilated convolution(s). Our experimental results on two publicly available polyp segmentation benchmark datasets showed that TransResU-Net obtained a highly promising dice score and a real-time speed. With high efficacy in our performance metrics, we concluded that TransResU-Net could be a strong benchmark for building a real-time polyp detection system for the early diagnosis, treatment, and prevention of colorectal cancer. The source code of the proposed TransResU-Net is publicly available at https://github.com/nikhilroxtomar/TransResUNet.
Partitioning signal classes using transport transforms for data analysis and machine learning
Aug 08, 2020



A relatively new set of transport-based transforms (CDT, R-CDT, LOT) have shown their strength and great potential in various image and data processing tasks such as parametric signal estimation, classification, cancer detection among many others. It is hence worthwhile to elucidate some of the mathematical properties that explain the successes of these transforms when they are used as tools in data analysis, signal processing or data classification. In particular, we give conditions under which classes of signals that are created by algebraic generative models are transformed into convex sets by the transport transforms. Such convexification of the classes simplify the classification and other data analysis and processing problems when viewed in the transform domain. More specifically, we study the extent and limitation of the convexification ability of these transforms under an algebraic generative modeling framework. We hope that this paper will serve as an introduction to these transforms and will encourage mathematicians and other researchers to further explore the theoretical underpinnings and algorithmic tools that will help understand the successes of these transforms and lay the groundwork for further successful applications.
Joint 2D-3D Breast Cancer Classification
Feb 27, 2020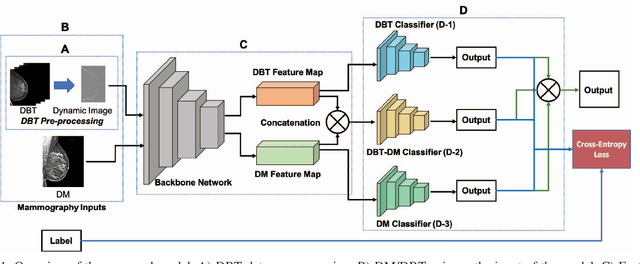
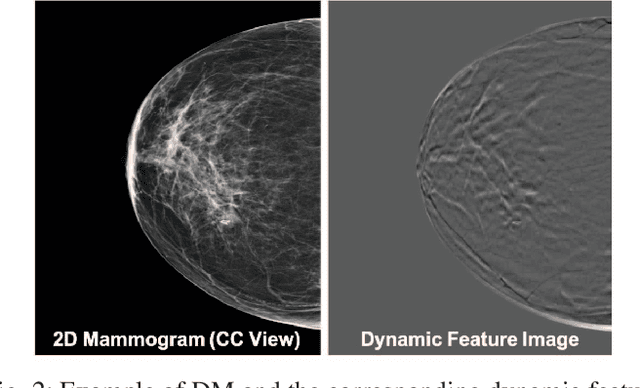
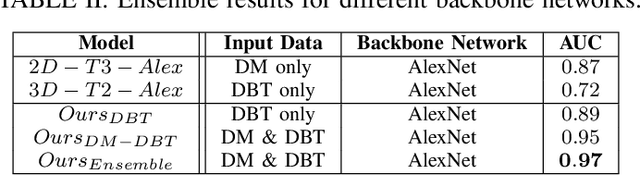
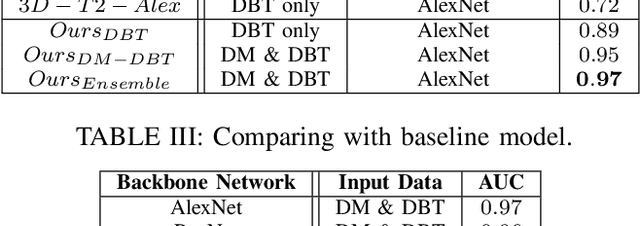
Breast cancer is the malignant tumor that causes the highest number of cancer deaths in females. Digital mammograms (DM or 2D mammogram) and digital breast tomosynthesis (DBT or 3D mammogram) are the two types of mammography imagery that are used in clinical practice for breast cancer detection and diagnosis. Radiologists usually read both imaging modalities in combination; however, existing computer-aided diagnosis tools are designed using only one imaging modality. Inspired by clinical practice, we propose an innovative convolutional neural network (CNN) architecture for breast cancer classification, which uses both 2D and 3D mammograms, simultaneously. Our experiment shows that the proposed method significantly improves the performance of breast cancer classification. By assembling three CNN classifiers, the proposed model achieves 0.97 AUC, which is 34.72% higher than the methods using only one imaging modality.
Semi-Supervised Learning for Cancer Detection of Lymph Node Metastases
Jun 23, 2019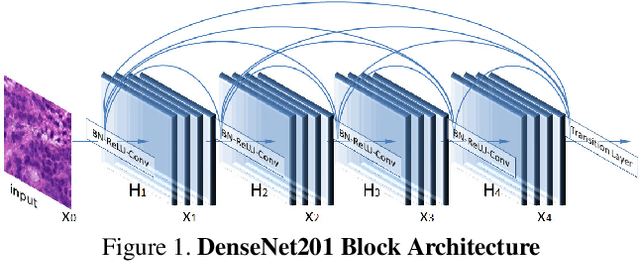
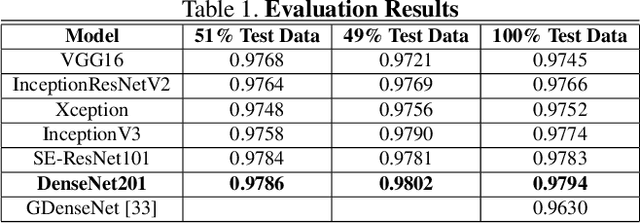
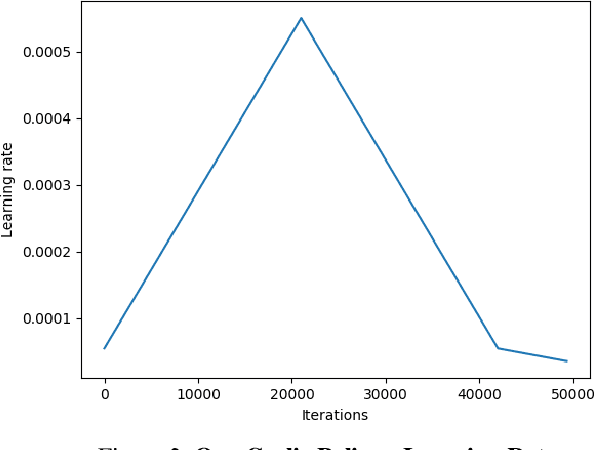

Pathologists find tedious to examine the status of the sentinel lymph node on a large number of pathological scans. The examination process of such lymph node which encompasses metastasized cancer cells is histopathologically organized. However, the task of finding metastatic tissues is gradual which is often challenging. In this work, we present our deep convolutional neural network based model validated on PatchCamelyon (PCam) benchmark dataset for fundamental machine learning research in histopathology diagnosis. We find that our proposed model trained with a semi-supervised learning approach by using pseudo labels on PCam-level significantly leads to better performances to strong CNN baseline on the AUC metric.
Colorectal cancer diagnosis from histology images: A comparative study
Mar 28, 2019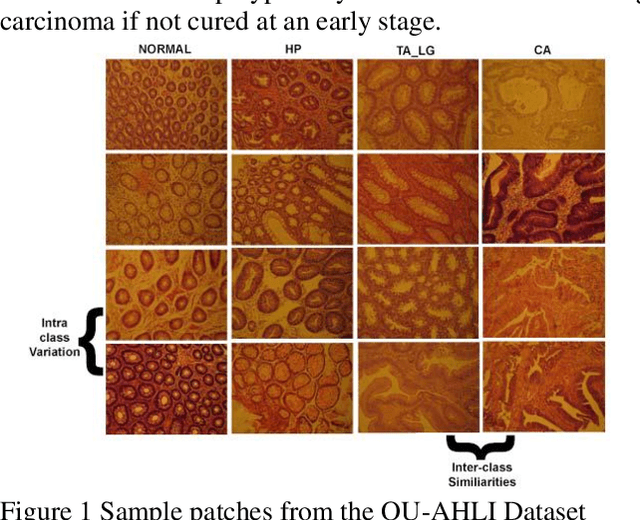
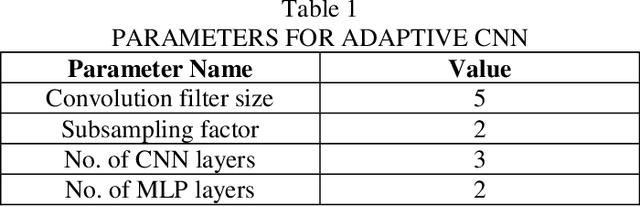
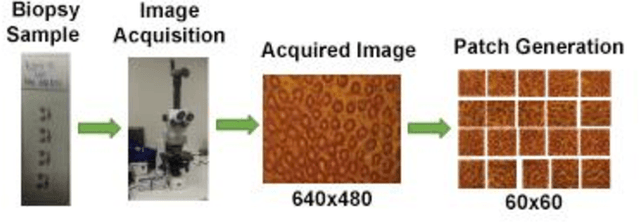
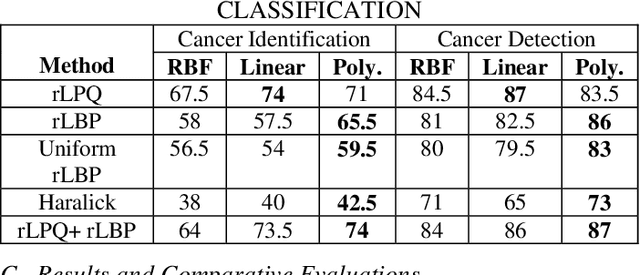
Computer-aided diagnosis (CAD) based on histopathological imaging has progressed rapidly in recent years with the rise of machine learning based methodologies. Traditional approaches consist of training a classification model using features extracted from the images, based on textures or morphological properties. Recently, deep-learning based methods have been applied directly to the raw (unprocessed) data. However, their usability is impacted by the paucity of annotated data in the biomedical sector. In order to leverage the learning capabilities of deep Convolutional Neural Nets (CNNs) within the confines of limited labelled data, in this study we shall investigate the transfer learning approaches that aim to apply the knowledge gained from solving a source (e.g., non-medical) problem, to learn better predictive models for the target (e.g., biomedical) task. As an alternative, we shall further propose a new adaptive and compact CNN based architecture that can be trained from scratch even on scarce and low-resolution data. Moreover, we conduct quantitative comparative evaluations among the traditional methods, transfer learning-based methods and the proposed adaptive approach for the particular task of cancer detection and identification from scarce and low-resolution histology images. Over the largest benchmark dataset formed for this purpose, the proposed adaptive approach achieved a higher cancer detection accuracy with a significant gap, whereas the deep CNNs with transfer learning achieved a superior cancer identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge