"cancer detection": models, code, and papers
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020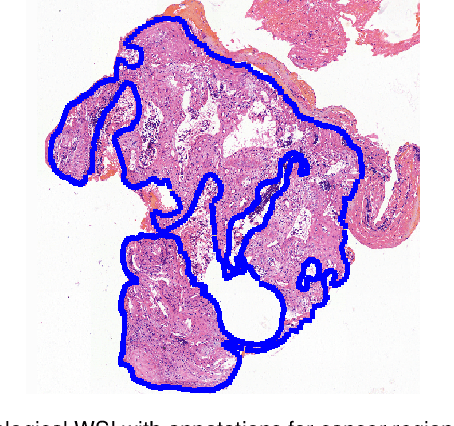
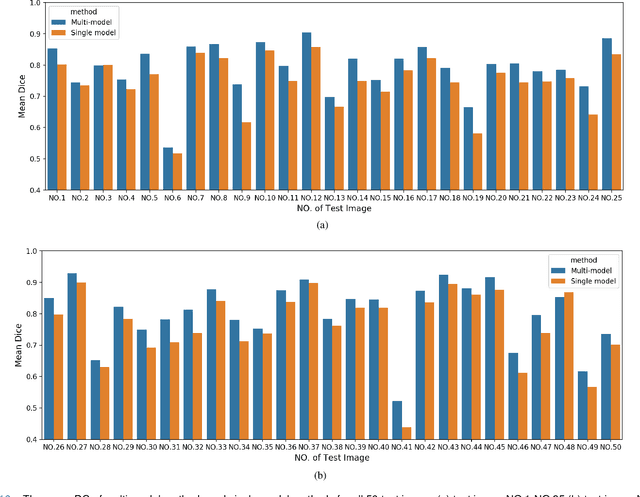
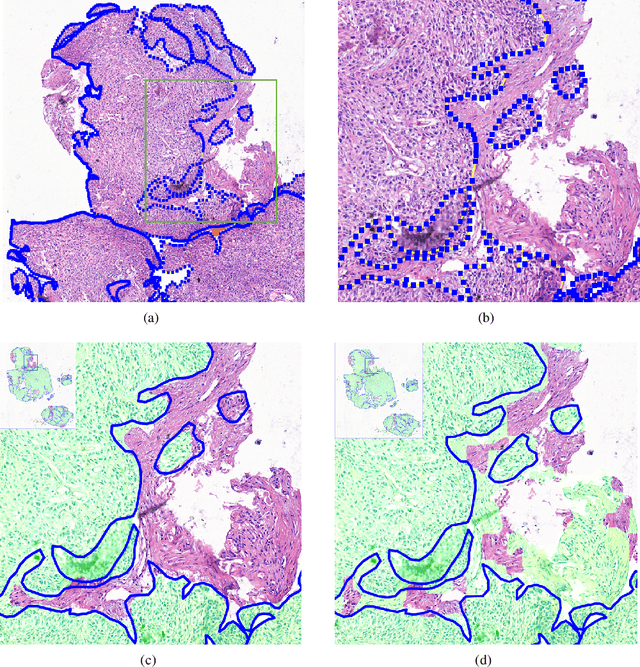
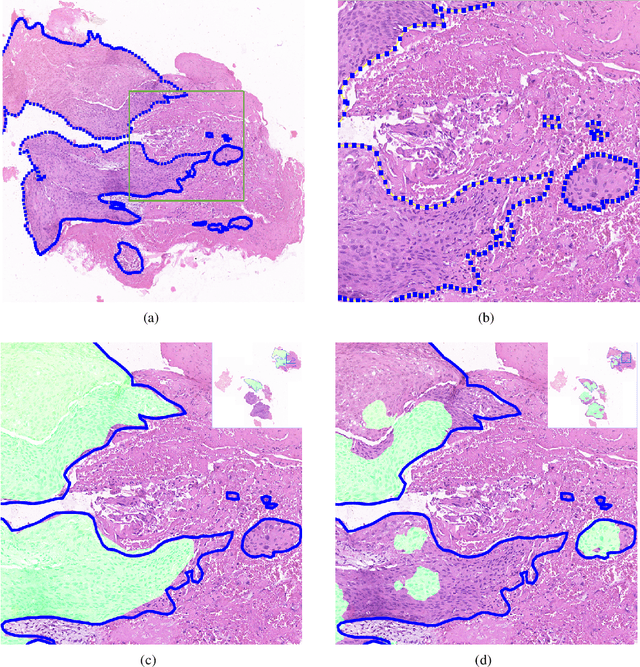
Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
Three Applications of Conformal Prediction for Rating Breast Density in Mammography
Jun 23, 2022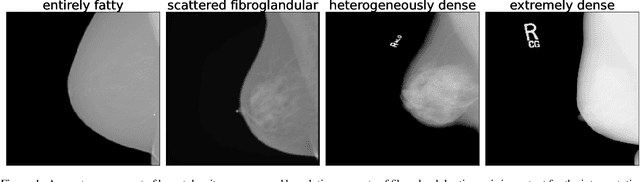

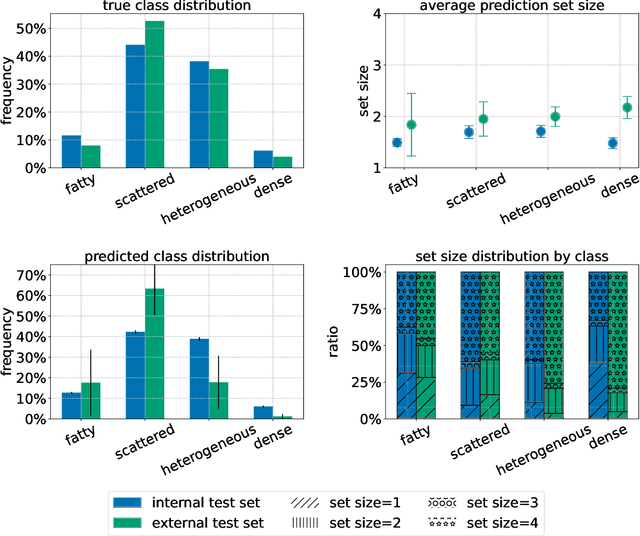
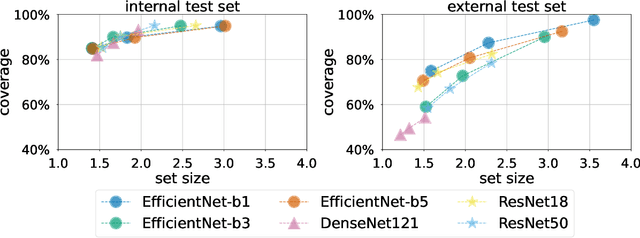
Breast cancer is the most common cancers and early detection from mammography screening is crucial in improving patient outcomes. Assessing mammographic breast density is clinically important as the denser breasts have higher risk and are more likely to occlude tumors. Manual assessment by experts is both time-consuming and subject to inter-rater variability. As such, there has been increased interest in the development of deep learning methods for mammographic breast density assessment. Despite deep learning having demonstrated impressive performance in several prediction tasks for applications in mammography, clinical deployment of deep learning systems in still relatively rare; historically, mammography Computer-Aided Diagnoses (CAD) have over-promised and failed to deliver. This is in part due to the inability to intuitively quantify uncertainty of the algorithm for the clinician, which would greatly enhance usability. Conformal prediction is well suited to increase reliably and trust in deep learning tools but they lack realistic evaluations on medical datasets. In this paper, we present a detailed analysis of three possible applications of conformal prediction applied to medical imaging tasks: distribution shift characterization, prediction quality improvement, and subgroup fairness analysis. Our results show the potential of distribution-free uncertainty quantification techniques to enhance trust on AI algorithms and expedite their translation to usage.
Random Forest as a Tumour Genetic Marker Extractor
Nov 26, 2019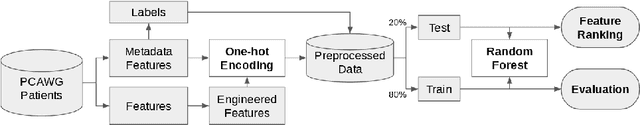
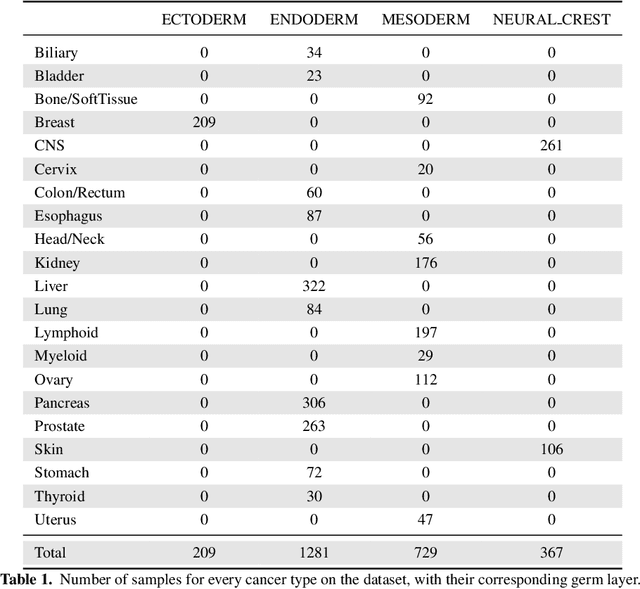
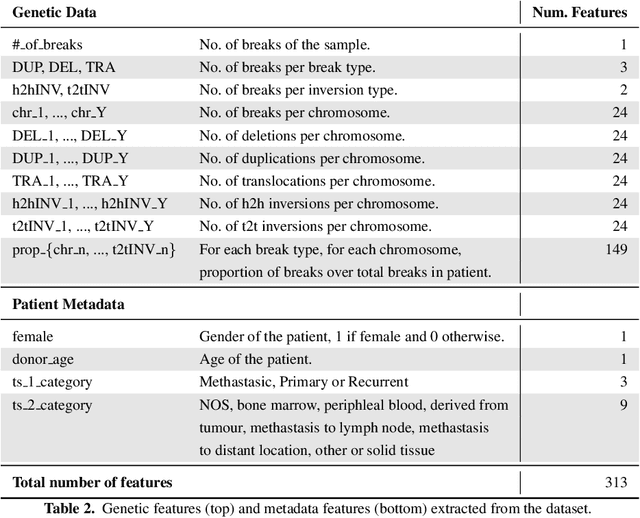
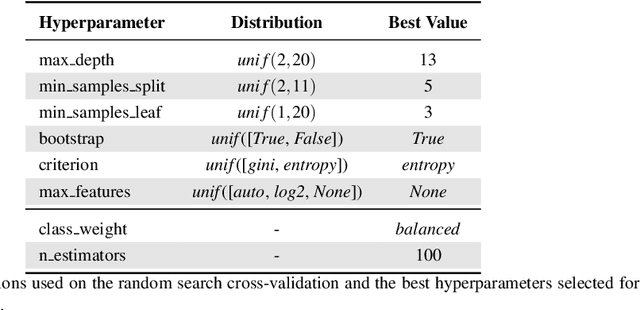
Finding tumour genetic markers is essential to biomedicine due to their relevance for cancer detection and therapy development. In this paper, we explore a recently released dataset of chromosome rearrangements in 2,586 cancer patients, where different sorts of alterations have been detected. Using a Random Forest classifier, we evaluate the relevance of several features (some directly available in the original data, some engineered by us) related to chromosome rearrangements. This evaluation results in a set of potential tumour genetic markers, some of which are validated in the bibliography, while others are potentially novel.
A New Dataset and A Baseline Model for Breast Lesion Detection in Ultrasound Videos
Jul 01, 2022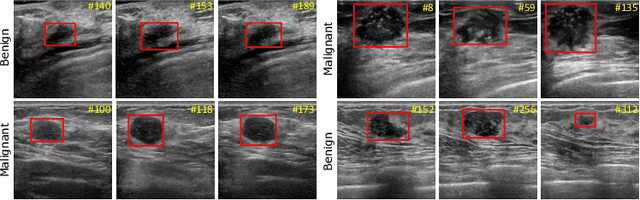
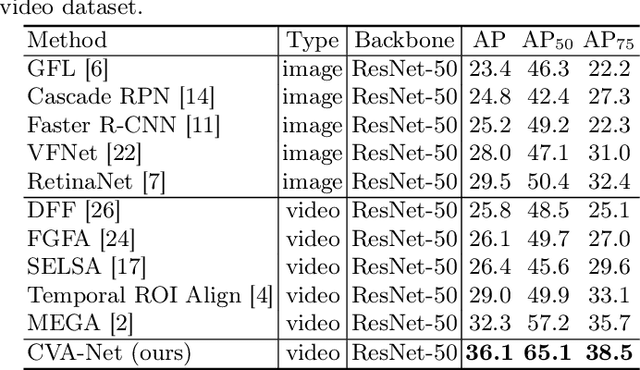
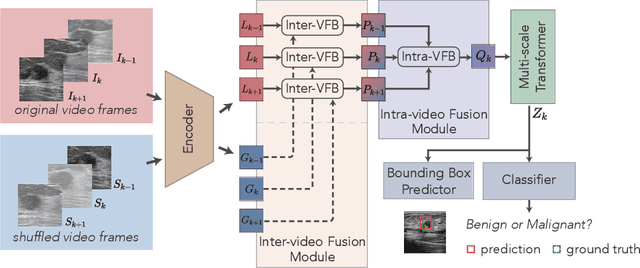
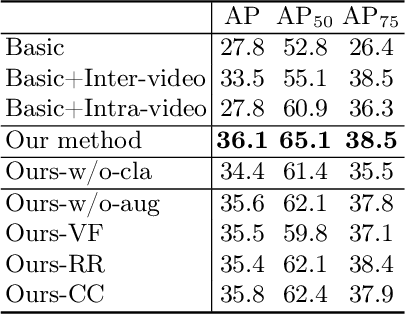
Breast lesion detection in ultrasound is critical for breast cancer diagnosis. Existing methods mainly rely on individual 2D ultrasound images or combine unlabeled video and labeled 2D images to train models for breast lesion detection. In this paper, we first collect and annotate an ultrasound video dataset (188 videos) for breast lesion detection. Moreover, we propose a clip-level and video-level feature aggregated network (CVA-Net) for addressing breast lesion detection in ultrasound videos by aggregating video-level lesion classification features and clip-level temporal features. The clip-level temporal features encode local temporal information of ordered video frames and global temporal information of shuffled video frames. In our CVA-Net, an inter-video fusion module is devised to fuse local features from original video frames and global features from shuffled video frames, and an intra-video fusion module is devised to learn the temporal information among adjacent video frames. Moreover, we learn video-level features to classify the breast lesions of the original video as benign or malignant lesions to further enhance the final breast lesion detection performance in ultrasound videos. Experimental results on our annotated dataset demonstrate that our CVA-Net clearly outperforms state-of-the-art methods. The corresponding code and dataset are publicly available at \url{https://github.com/jhl-Det/CVA-Net}.
* 11 pages, 4 figures
EBHI:A New Enteroscope Biopsy Histopathological H&E Image Dataset for Image Classification Evaluation
Feb 17, 2022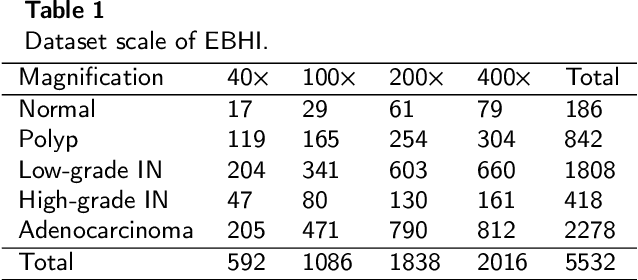
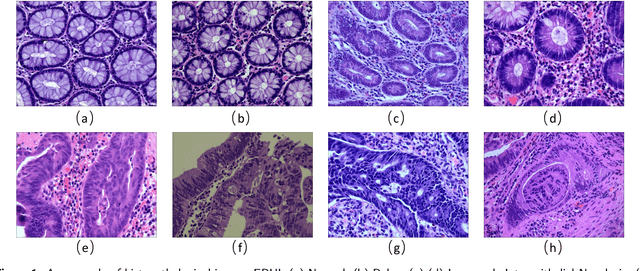
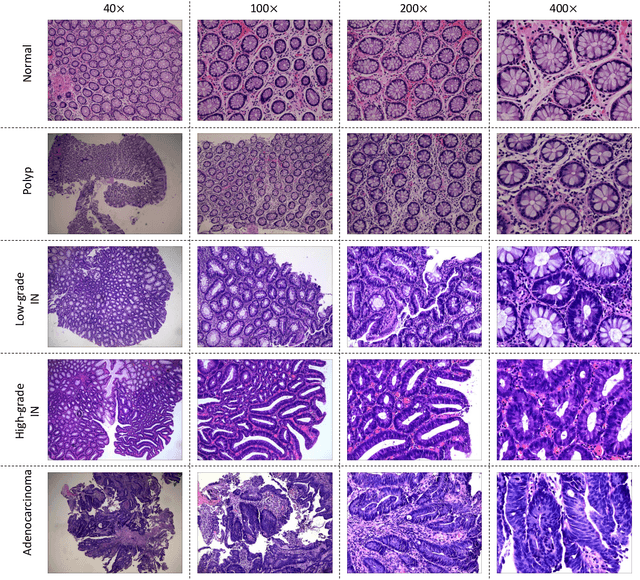
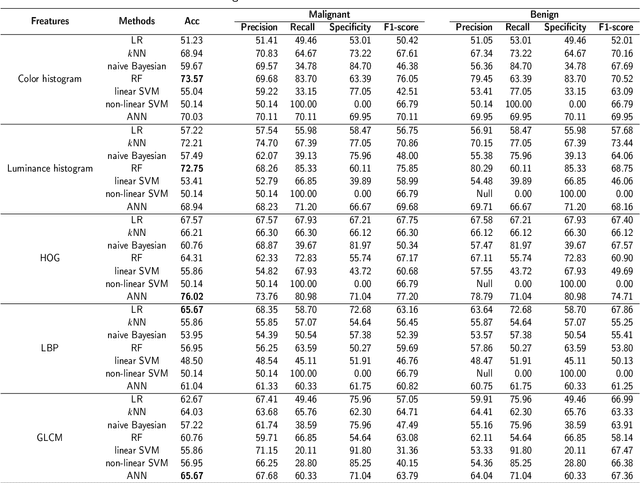
Background and purpose: Colorectal cancer has become the third most common cancer worldwide, accounting for approximately 10% of cancer patients. Early detection of the disease is important for the treatment of colorectal cancer patients. Histopathological examination is the gold standard for screening colorectal cancer. However, the current lack of histopathological image datasets of colorectal cancer, especially enteroscope biopsies, hinders the accurate evaluation of computer-aided diagnosis techniques. Methods: A new publicly available Enteroscope Biopsy Histopathological H&E Image Dataset (EBHI) is published in this paper. To demonstrate the effectiveness of the EBHI dataset, we have utilized several machine learning, convolutional neural networks and novel transformer-based classifiers for experimentation and evaluation, using an image with a magnification of 200x. Results: Experimental results show that the deep learning method performs well on the EBHI dataset. Traditional machine learning methods achieve maximum accuracy of 76.02% and deep learning method achieves a maximum accuracy of 95.37%. Conclusion: To the best of our knowledge, EBHI is the first publicly available colorectal histopathology enteroscope biopsy dataset with four magnifications and five types of images of tumor differentiation stages, totaling 5532 images. We believe that EBHI could attract researchers to explore new classification algorithms for the automated diagnosis of colorectal cancer, which could help physicians and patients in clinical settings.
Sharing Generative Models Instead of Private Data: A Simulation Study on Mammography Patch Classification
Mar 08, 2022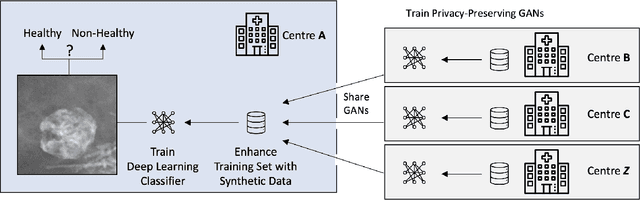
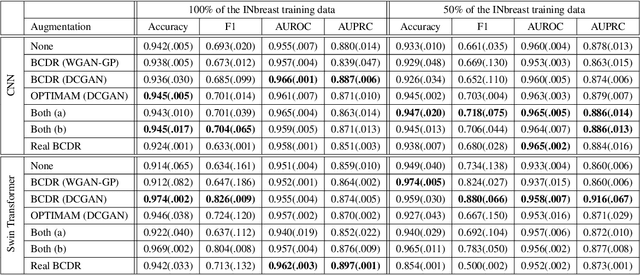
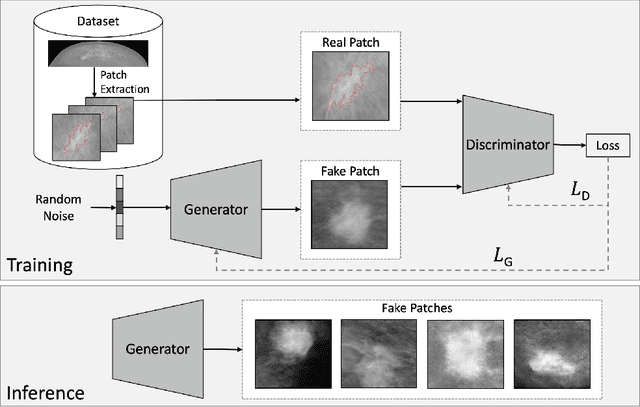
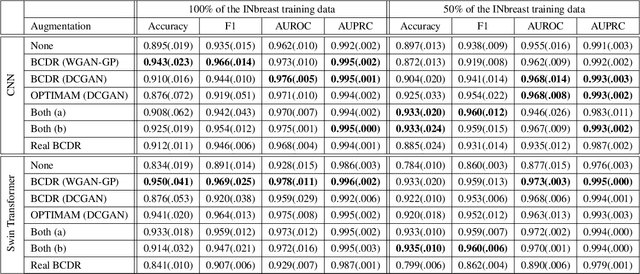
Early detection of breast cancer in mammography screening via deep-learning based computer-aided detection systems shows promising potential in improving the curability and mortality rates of breast cancer. However, many clinical centres are restricted in the amount and heterogeneity of available data to train such models to (i) achieve promising performance and to (ii) generalise well across acquisition protocols and domains. As sharing data between centres is restricted due to patient privacy concerns, we propose a potential solution: sharing trained generative models between centres as substitute for real patient data. In this work, we use three well known mammography datasets to simulate three different centres, where one centre receives the trained generator of Generative Adversarial Networks (GANs) from the two remaining centres in order to augment the size and heterogeneity of its training dataset. We evaluate the utility of this approach on mammography patch classification on the test set of the GAN-receiving centre using two different classification models, (a) a convolutional neural network and (b) a transformer neural network. Our experiments demonstrate that shared GANs notably increase the performance of both transformer and convolutional classification models and highlight this approach as a viable alternative to inter-centre data sharing.
Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels
Jun 05, 2020
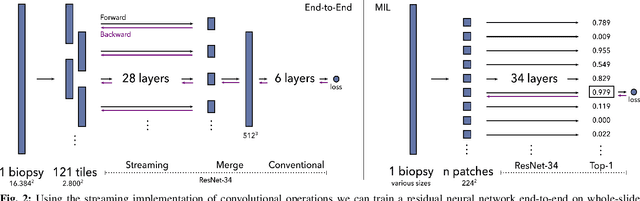
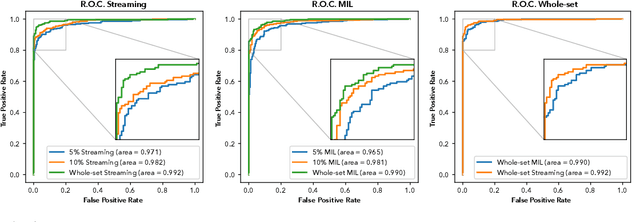

Prostate cancer is the most prevalent cancer among men in Western countries, with 1.1 million new diagnoses every year. The gold standard for the diagnosis of prostate cancer is a pathologists' evaluation of prostate tissue. To potentially assist pathologists deep-learning-based cancer detection systems have been developed. Many of the state-of-the-art models are patch-based convolutional neural networks, as the use of entire scanned slides is hampered by memory limitations on accelerator cards. Patch-based systems typically require detailed, pixel-level annotations for effective training. However, such annotations are seldom readily available, in contrast to the clinical reports of pathologists, which contain slide-level labels. As such, developing algorithms which do not require manual pixel-wise annotations, but can learn using only the clinical report would be a significant advancement for the field. In this paper, we propose to use a streaming implementation of convolutional layers, to train a modern CNN (ResNet-34) with 21 million parameters end-to-end on 4712 prostate biopsies. The method enables the use of entire biopsy images at high-resolution directly by reducing the GPU memory requirements by 2.4 TB. We show that modern CNNs, trained using our streaming approach, can extract meaningful features from high-resolution images without additional heuristics, reaching similar performance as state-of-the-art patch-based and multiple-instance learning methods. By circumventing the need for manual annotations, this approach can function as a blueprint for other tasks in histopathological diagnosis. The source code to reproduce the streaming models is available at https://github.com/DIAGNijmegen/pathology-streaming-pipeline .
One-Pixel Attack Deceives Automatic Detection of Breast Cancer
Dec 16, 2020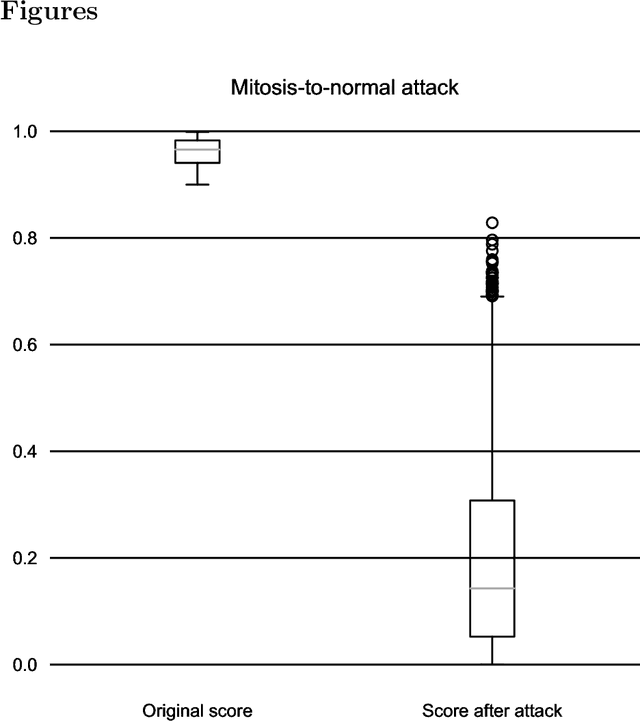
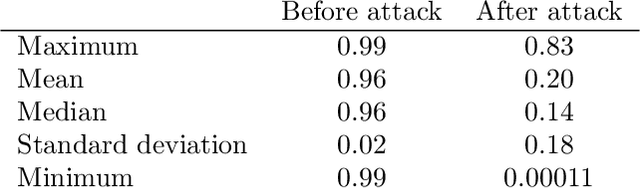
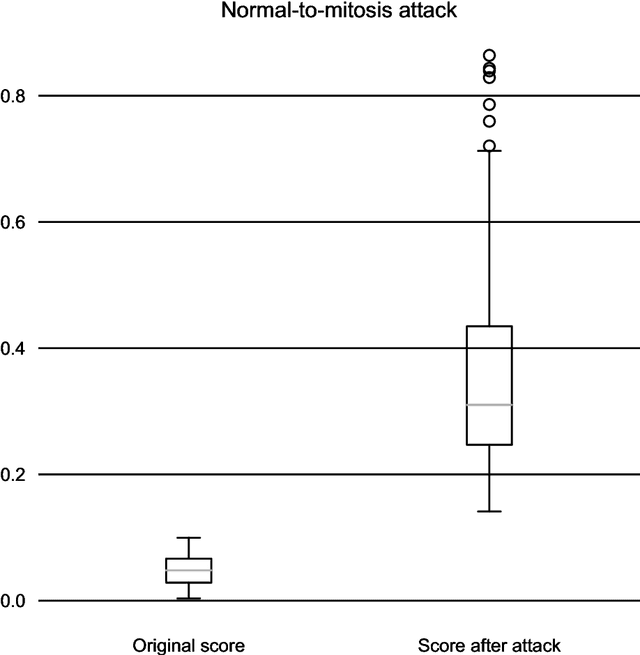
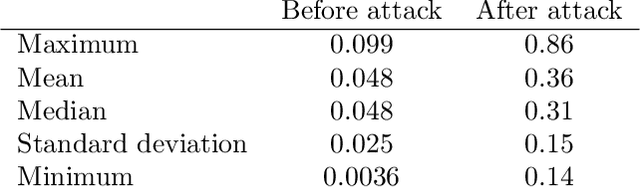
In this article we demonstrate that a state-of-the-art machine learning model predicting whether a whole slide image contains mitosis can be fooled by changing just a single pixel in the input image. Computer vision and machine learning can be used to automate various tasks in cancer diagnostic and detection. If an attacker can manipulate the automated processing, the results can be devastating and in the worst case lead to wrong diagnostic and treatments. In this research one-pixel attack is demonstrated in a real-life scenario with a real tumor dataset. The results indicate that a minor one-pixel modification of a whole slide image under analysis can affect the diagnosis. The attack poses a threat from the cyber security perspective: the one-pixel method can be used as an attack vector by a motivated attacker.
Unsupervised Learning of Deep-Learned Features from Breast Cancer Images
Jun 21, 2020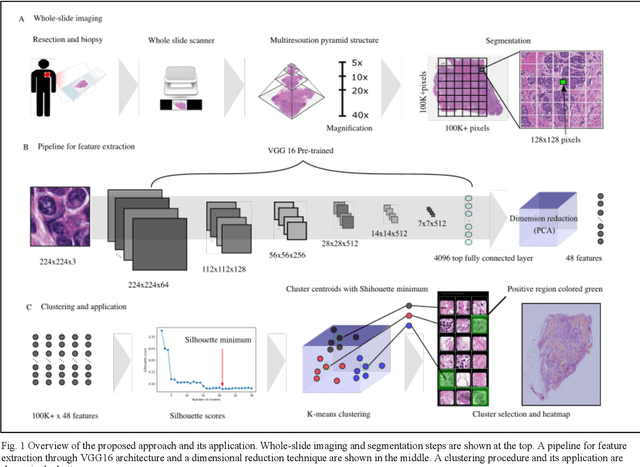
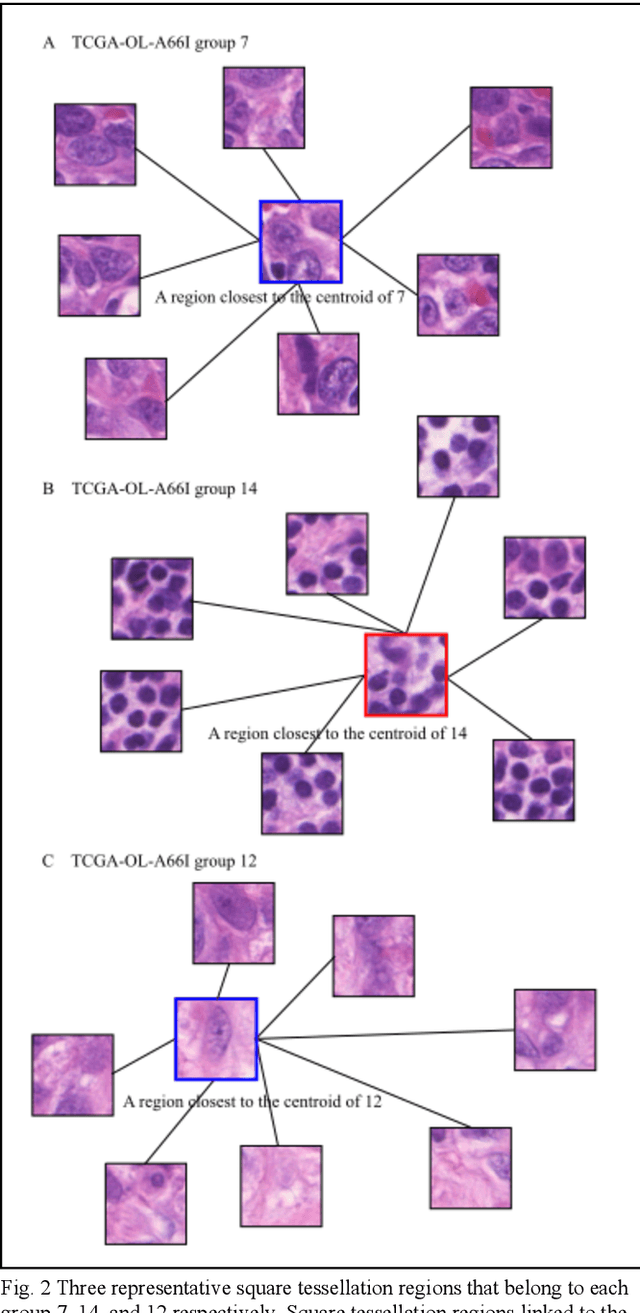
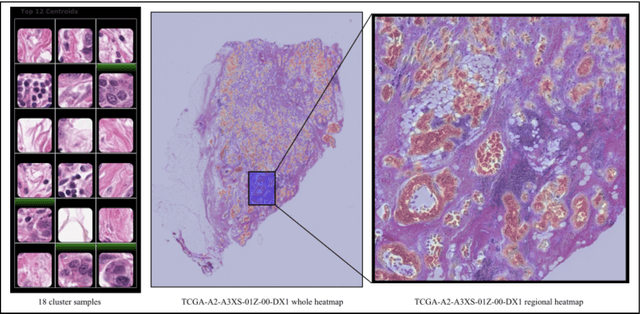
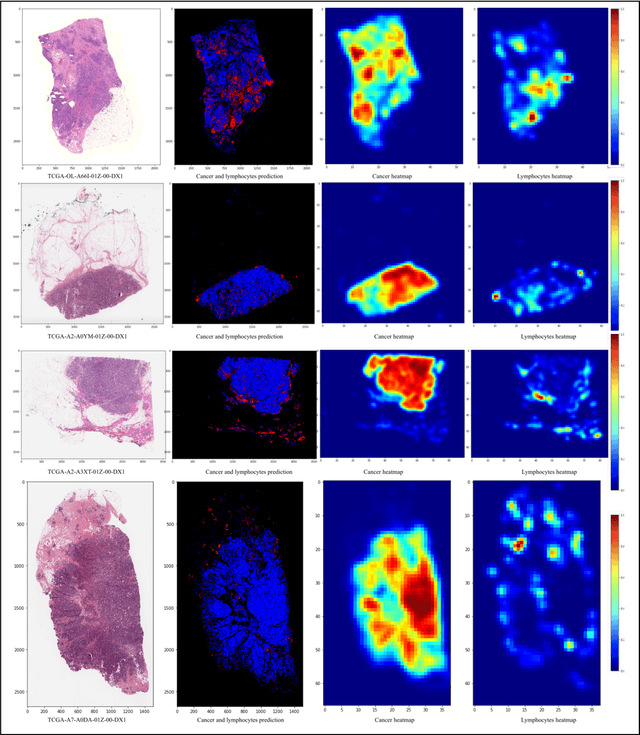
Detecting cancer manually in whole slide images requires significant time and effort on the laborious process. Recent advances in whole slide image analysis have stimulated the growth and development of machine learning-based approaches that improve the efficiency and effectiveness in the diagnosis of cancer diseases. In this paper, we propose an unsupervised learning approach for detecting cancer in breast invasive carcinoma (BRCA) whole slide images. The proposed method is fully automated and does not require human involvement during the unsupervised learning procedure. We demonstrate the effectiveness of the proposed approach for cancer detection in BRCA and show how the machine can choose the most appropriate clusters during the unsupervised learning procedure. Moreover, we present a prototype application that enables users to select relevant groups mapping all regions related to the groups in whole slide images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge