"cancer detection": models, code, and papers
Surpassing the Human Accuracy: Detecting Gallbladder Cancer from USG Images with Curriculum Learning
Apr 25, 2022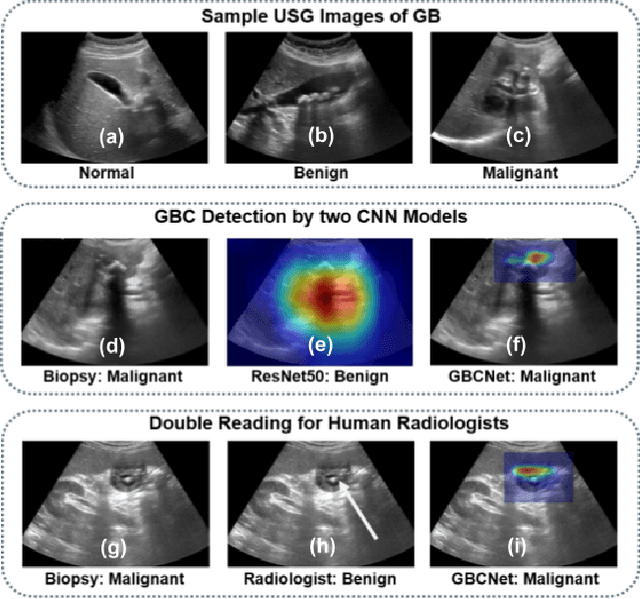
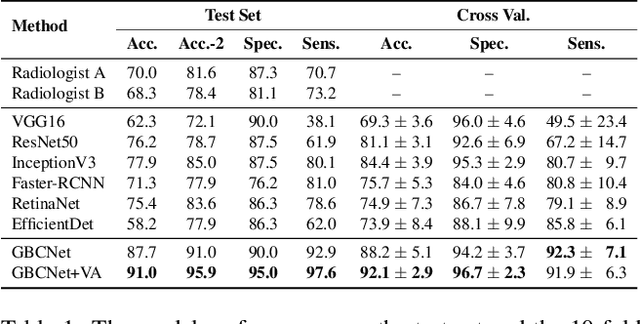
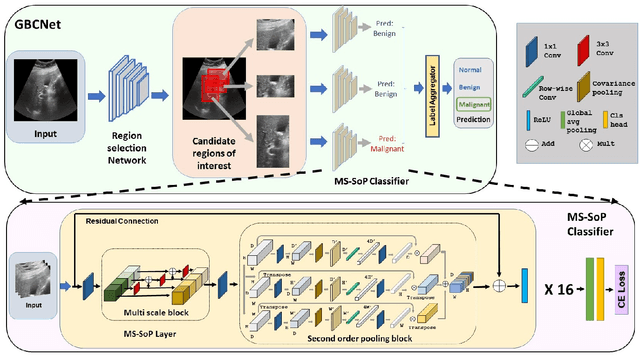
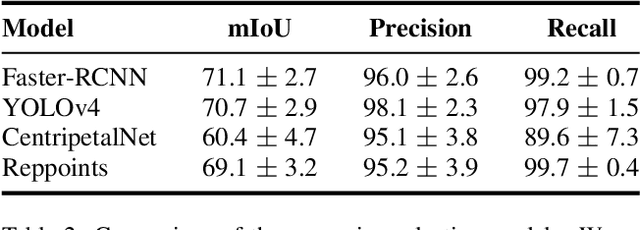
We explore the potential of CNN-based models for gallbladder cancer (GBC) detection from ultrasound (USG) images as no prior study is known. USG is the most common diagnostic modality for GB diseases due to its low cost and accessibility. However, USG images are challenging to analyze due to low image quality, noise, and varying viewpoints due to the handheld nature of the sensor. Our exhaustive study of state-of-the-art (SOTA) image classification techniques for the problem reveals that they often fail to learn the salient GB region due to the presence of shadows in the USG images. SOTA object detection techniques also achieve low accuracy because of spurious textures due to noise or adjacent organs. We propose GBCNet to tackle the challenges in our problem. GBCNet first extracts the regions of interest (ROIs) by detecting the GB (and not the cancer), and then uses a new multi-scale, second-order pooling architecture specializing in classifying GBC. To effectively handle spurious textures, we propose a curriculum inspired by human visual acuity, which reduces the texture biases in GBCNet. Experimental results demonstrate that GBCNet significantly outperforms SOTA CNN models, as well as the expert radiologists. Our technical innovations are generic to other USG image analysis tasks as well. Hence, as a validation, we also show the efficacy of GBCNet in detecting breast cancer from USG images. Project page with source code, trained models, and data is available at https://gbc-iitd.github.io/gbcnet
Lung Cancer Detection using Co-learning from Chest CT Images and Clinical Demographics
Feb 21, 2019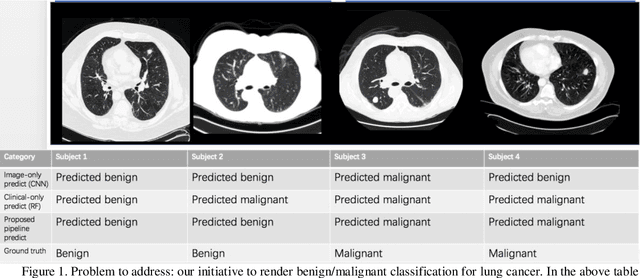
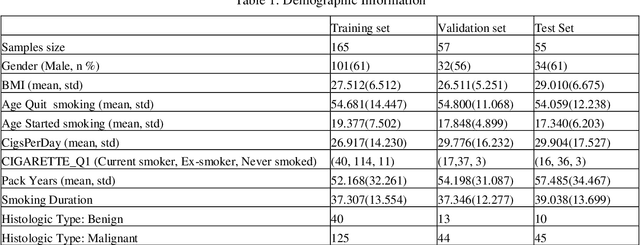
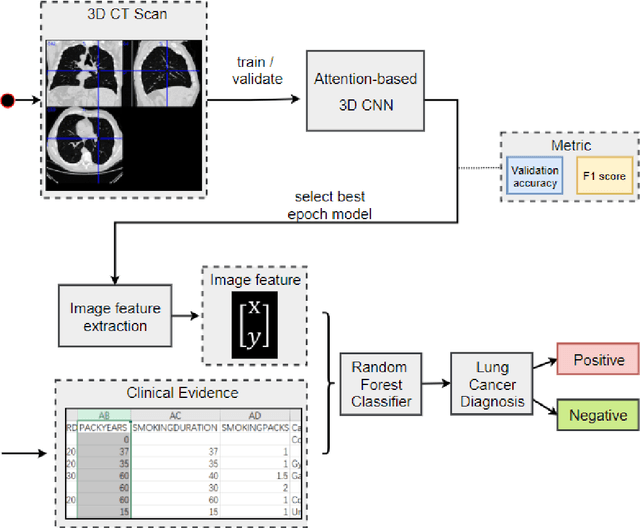
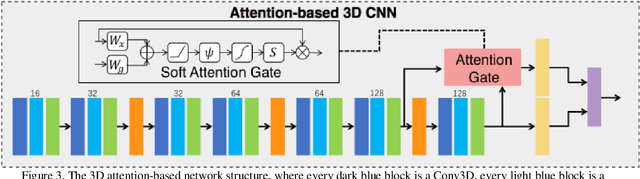
Early detection of lung cancer is essential in reducing mortality. Recent studies have demonstrated the clinical utility of low-dose computed tomography (CT) to detect lung cancer among individuals selected based on very limited clinical information. However, this strategy yields high false positive rates, which can lead to unnecessary and potentially harmful procedures. To address such challenges, we established a pipeline that co-learns from detailed clinical demographics and 3D CT images. Toward this end, we leveraged data from the Consortium for Molecular and Cellular Characterization of Screen-Detected Lesions (MCL), which focuses on early detection of lung cancer. A 3D attention-based deep convolutional neural net (DCNN) is proposed to identify lung cancer from the chest CT scan without prior anatomical location of the suspicious nodule. To improve upon the non-invasive discrimination between benign and malignant, we applied a random forest classifier to a dataset integrating clinical information to imaging data. The results show that the AUC obtained from clinical demographics alone was 0.635 while the attention network alone reached an accuracy of 0.687. In contrast when applying our proposed pipeline integrating clinical and imaging variables, we reached an AUC of 0.787 on the testing dataset. The proposed network both efficiently captures anatomical information for classification and also generates attention maps that explain the features that drive performance.
Demystifying the Transferability of Adversarial Attacks in Computer Networks
Oct 09, 2021
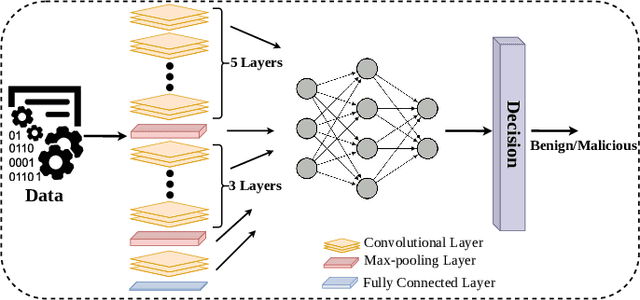
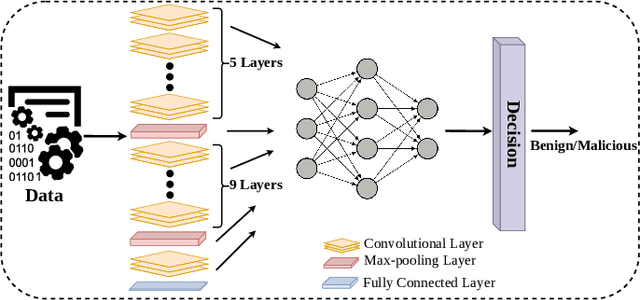
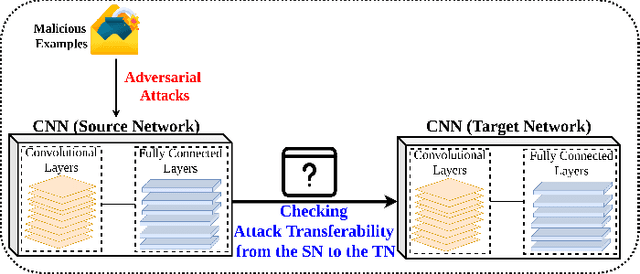
Deep Convolutional Neural Networks (CNN) models are one of the most popular networks in deep learning. With their large fields of application in different areas, they are extensively used in both academia and industry. CNN-based models include several exciting implementations such as early breast cancer detection or detecting developmental delays in children (e.g., autism, speech disorders, etc.). However, previous studies demonstrate that these models are subject to various adversarial attacks. Interestingly, some adversarial examples could potentially still be effective against different unknown models. This particular property is known as adversarial transferability, and prior works slightly analyzed this characteristic in a very limited application domain. In this paper, we aim to demystify the transferability threats in computer networks by studying the possibility of transferring adversarial examples. In particular, we provide the first comprehensive study which assesses the robustness of CNN-based models for computer networks against adversarial transferability. In our experiments, we consider five different attacks: (1) the Iterative Fast Gradient Method (I-FGSM), (2) the Jacobian-based Saliency Map attack (JSMA), (3) the L-BFGS attack, (4) the Projected Gradient Descent attack (PGD), and (5) the DeepFool attack. These attacks are performed against two well-known datasets: the N-BaIoT dataset and the Domain Generating Algorithms (DGA) dataset. Our results show that the transferability happens in specific use cases where the adversary can easily compromise the victim's network with very few knowledge of the targeted model.
Wide & Deep neural network model for patch aggregation in CNN-based prostate cancer detection systems
May 20, 2021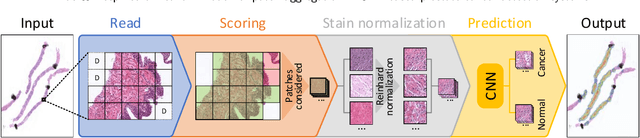
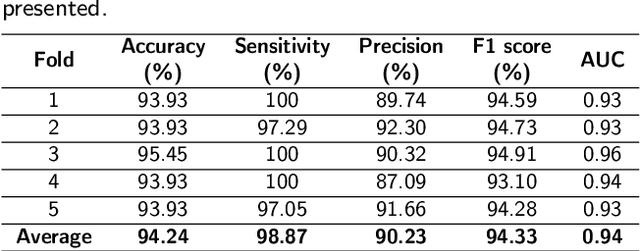
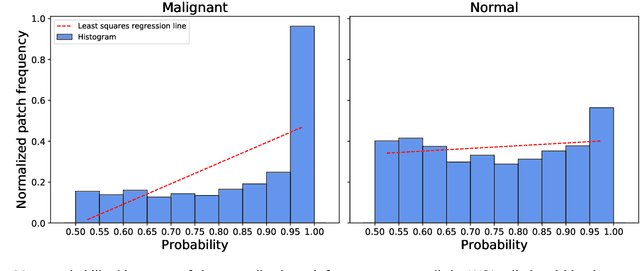
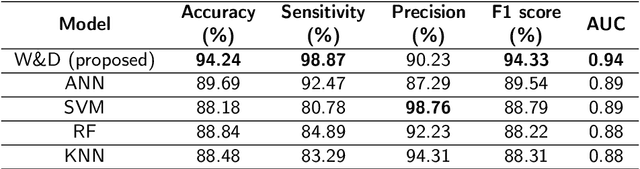
Prostate cancer (PCa) is one of the most commonly diagnosed cancer and one of the leading causes of death among men, with almost 1.41 million new cases and around 375,000 deaths in 2020. Artificial Intelligence algorithms have had a huge impact in medical image analysis, including digital histopathology, where Convolutional Neural Networks (CNNs) are used to provide a fast and accurate diagnosis, supporting experts in this task. To perform an automatic diagnosis, prostate tissue samples are first digitized into gigapixel-resolution whole-slide images. Due to the size of these images, neural networks cannot use them as input and, therefore, small subimages called patches are extracted and predicted, obtaining a patch-level classification. In this work, a novel patch aggregation method based on a custom Wide & Deep neural network model is presented, which performs a slide-level classification using the patch-level classes obtained from a CNN. The malignant tissue ratio, a 10-bin malignant probability histogram, the least squares regression line of the histogram, and the number of malignant connected components are used by the proposed model to perform the classification. An accuracy of 94.24% and a sensitivity of 98.87% were achieved, proving that the proposed system could aid pathologists by speeding up the screening process and, thus, contribute to the fight against PCa.
FCN-Transformer Feature Fusion for Polyp Segmentation
Aug 17, 2022Colonoscopy is widely recognised as the gold standard procedure for the early detection of colorectal cancer (CRC). Segmentation is valuable for two significant clinical applications, namely lesion detection and classification, providing means to improve accuracy and robustness. The manual segmentation of polyps in colonoscopy images is time-consuming. As a result, the use of deep learning (DL) for automation of polyp segmentation has become important. However, DL-based solutions can be vulnerable to overfitting and the resulting inability to generalise to images captured by different colonoscopes. Recent transformer-based architectures for semantic segmentation both achieve higher performance and generalise better than alternatives, however typically predict a segmentation map of $\frac{h}{4}\times\frac{w}{4}$ spatial dimensions for a $h\times w$ input image. To this end, we propose a new architecture for full-size segmentation which leverages the strengths of a transformer in extracting the most important features for segmentation in a primary branch, while compensating for its limitations in full-size prediction with a secondary fully convolutional branch. The resulting features from both branches are then fused for final prediction of a $h\times w$ segmentation map. We demonstrate our method's state-of-the-art performance with respect to the mDice, mIoU, mPrecision, and mRecall metrics, on both the Kvasir-SEG and CVC-ClinicDB dataset benchmarks. Additionally, we train the model on each of these datasets and evaluate on the other to demonstrate its superior generalisation performance.
* 16 pages, 4 figures
Internal-transfer Weighting of Multi-task Learning for Lung Cancer Detection
Dec 16, 2019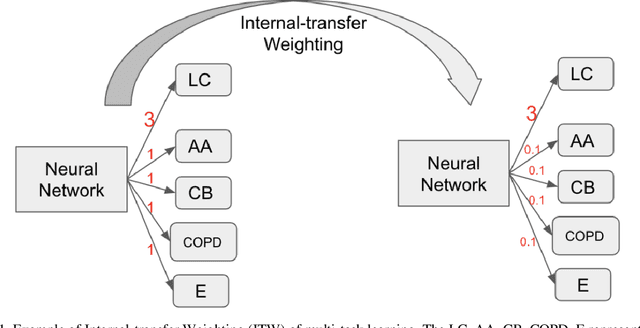
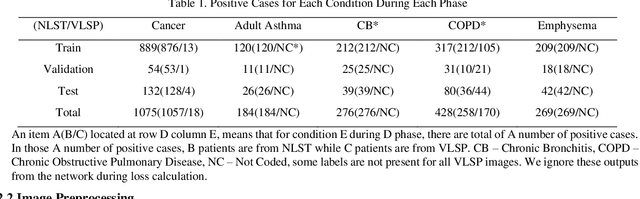
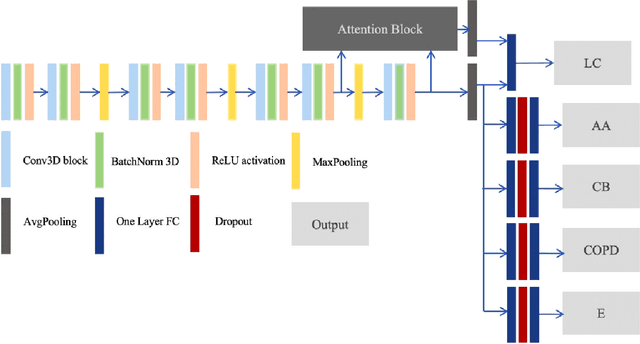
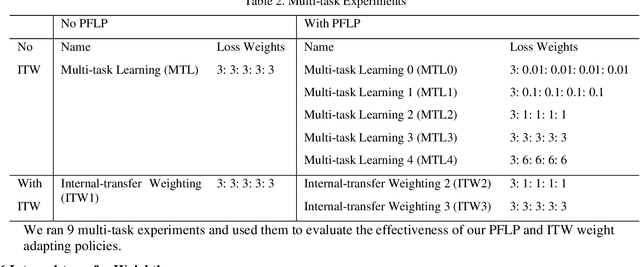
Recently, multi-task networks have shown to both offer additional estimation capabilities, and, perhaps more importantly, increased performance over single-task networks on a "main/primary" task. However, balancing the optimization criteria of multi-task networks across different tasks is an area of active exploration. Here, we extend a previously proposed 3D attention-based network with four additional multi-task subnetworks for the detection of lung cancer and four auxiliary tasks (diagnosis of asthma, chronic bronchitis, chronic obstructive pulmonary disease, and emphysema). We introduce and evaluate a learning policy, Periodic Focusing Learning Policy (PFLP), that alternates the dominance of tasks throughout the training. To improve performance on the primary task, we propose an Internal-Transfer Weighting (ITW) strategy to suppress the loss functions on auxiliary tasks for the final stages of training. To evaluate this approach, we examined 3386 patients (single scan per patient) from the National Lung Screening Trial (NLST) and de-identified data from the Vanderbilt Lung Screening Program, with a 2517/277/592 (scans) split for training, validation, and testing. Baseline networks include a single-task strategy and a multi-task strategy without adaptive weights (PFLP/ITW), while primary experiments are multi-task trials with either PFLP or ITW or both. On the test set for lung cancer prediction, the baseline single-task network achieved prediction AUC of 0.8080 and the multi-task baseline failed to converge (AUC 0.6720). However, applying PFLP helped multi-task network clarify and achieved test set lung cancer prediction AUC of 0.8402. Furthermore, our ITW technique boosted the PFLP enabled multi-task network and achieved an AUC of 0.8462 (McNemar test, p < 0.01).
Hybrid Machine Learning Model of Extreme Learning Machine Radial basis function for Breast Cancer Detection and Diagnosis; a Multilayer Fuzzy Expert System
Oct 29, 2019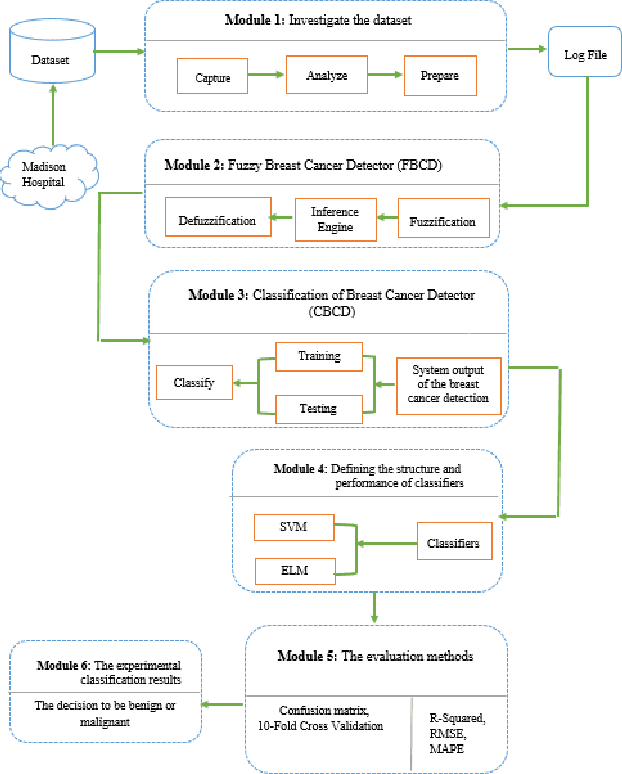
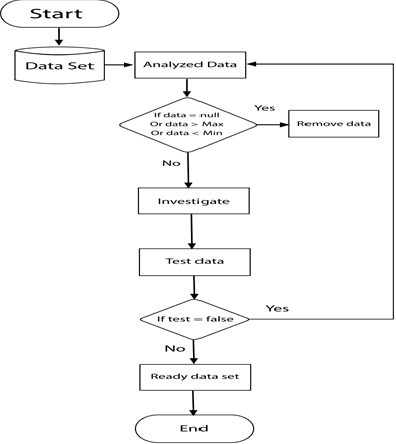
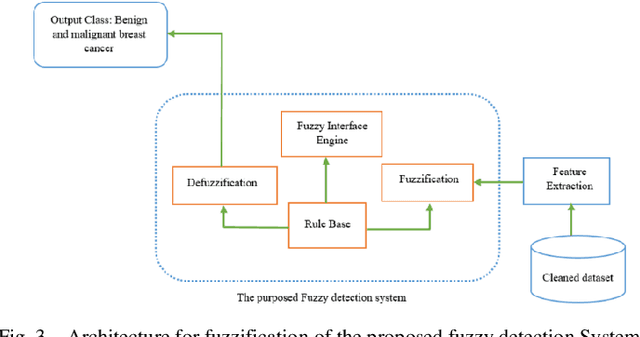
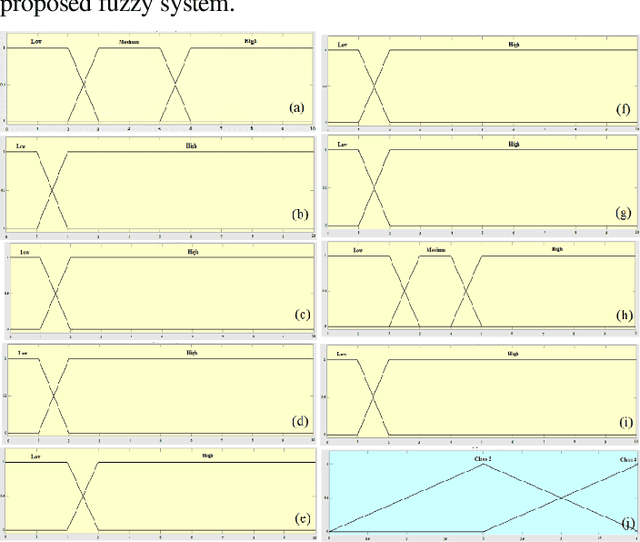
Mammography is often used as the most common laboratory method for the detection of breast cancer, yet associated with the high cost and many side effects. Machine learning prediction as an alternative method has shown promising results. This paper presents a method based on a multilayer fuzzy expert system for the detection of breast cancer using an extreme learning machine (ELM) classification model integrated with radial basis function (RBF) kernel called ELM-RBF, considering the Wisconsin dataset. The performance of the proposed model is further compared with a linear-SVM model. The proposed model outperforms the linear-SVM model with RMSE, R2, MAPE equal to 0.1719, 0.9374 and 0.0539, respectively. Furthermore, both models are studied in terms of criteria of accuracy, precision, sensitivity, specificity, validation, true positive rate (TPR), and false-negative rate (FNR). The ELM-RBF model for these criteria presents better performance compared to the SVM model.
Controlling False Positive/Negative Rates for Deep-Learning-Based Prostate Cancer Detection on Multiparametric MR images
Jun 04, 2021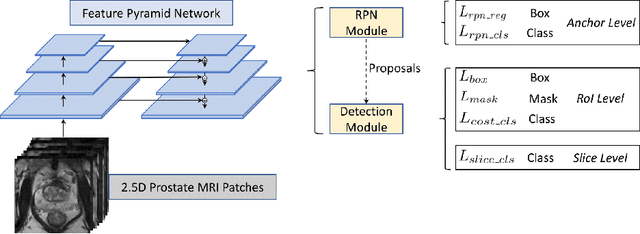

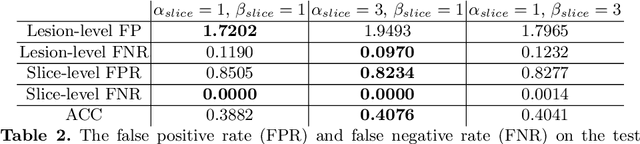
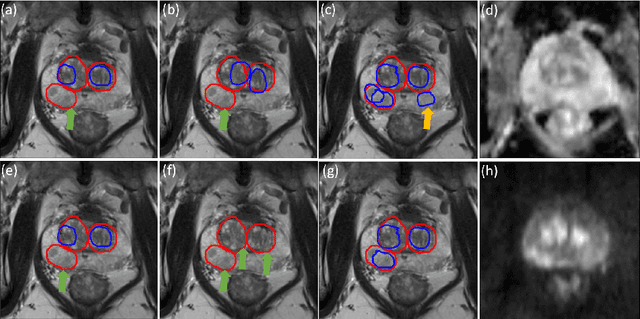
Prostate cancer (PCa) is one of the leading causes of death for men worldwide. Multi-parametric magnetic resonance (mpMR) imaging has emerged as a non-invasive diagnostic tool for detecting and localising prostate tumours by specialised radiologists. These radiological examinations, for example, for differentiating malignant lesions from benign prostatic hyperplasia in transition zones and for defining the boundaries of clinically significant cancer, remain challenging and highly skill-and-experience-dependent. We first investigate experimental results in developing object detection neural networks that are trained to predict the radiological assessment, using these high-variance labels. We further argue that such a computer-assisted diagnosis (CAD) system needs to have the ability to control the false-positive rate (FPR) or false-negative rate (FNR), in order to be usefully deployed in a clinical workflow, informing clinical decisions without further human intervention. This work proposes a novel PCa detection network that incorporates a lesion-level cost-sensitive loss and an additional slice-level loss based on a lesion-to-slice mapping function, to manage the lesion- and slice-level costs, respectively. Our experiments based on 290 clinical patients concludes that 1) The lesion-level FNR was effectively reduced from 0.19 to 0.10 and the lesion-level FPR was reduced from 1.03 to 0.66 by changing the lesion-level cost; 2) The slice-level FNR was reduced from 0.19 to 0.00 by taking into account the slice-level cost; (3) Both lesion-level and slice-level FNRs were reduced with lower FP/FPR by changing the lesion-level or slice-level costs, compared with post-training threshold adjustment using networks without the proposed cost-aware training.
Adversarial Networks for Prostate Cancer Detection
Nov 28, 2017
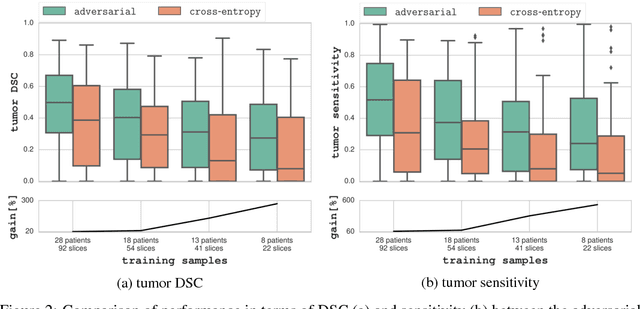
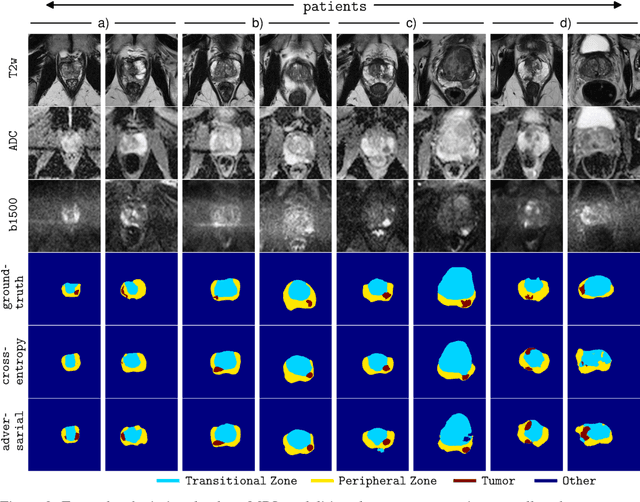
The large number of trainable parameters of deep neural networks renders them inherently data hungry. This characteristic heavily challenges the medical imaging community and to make things even worse, many imaging modalities are ambiguous in nature leading to rater-dependant annotations that current loss formulations fail to capture. We propose employing adversarial training for segmentation networks in order to alleviate aforementioned problems. We learn to segment aggressive prostate cancer utilizing challenging MRI images of 152 patients and show that the proposed scheme is superior over the de facto standard in terms of the detection sensitivity and the dice-score for aggressive prostate cancer. The achieved relative gains are shown to be particularly pronounced in the small dataset limit.
An Enhanced Deep Learning Technique for Prostate Cancer Identification Based on MRI Scans
Aug 01, 2022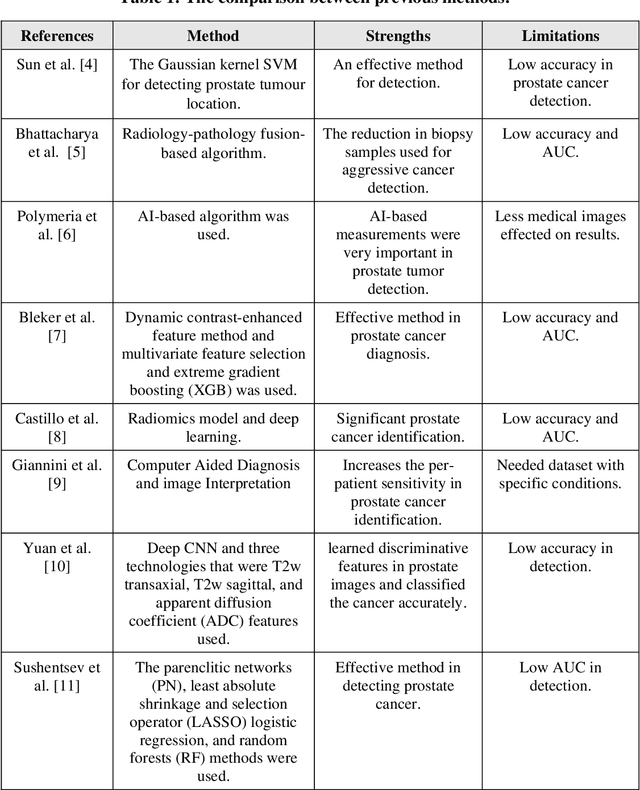
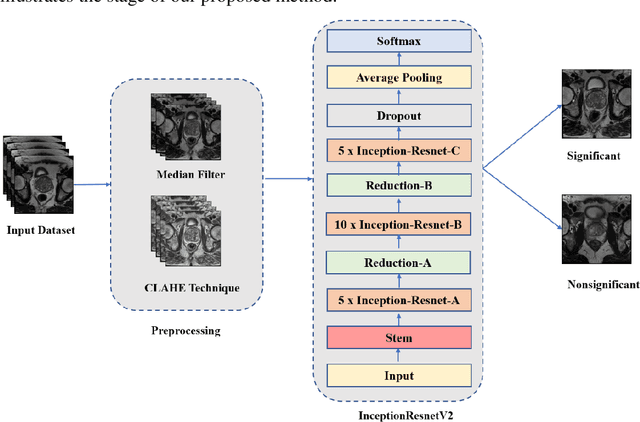
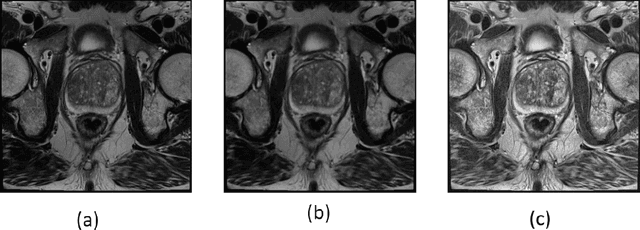
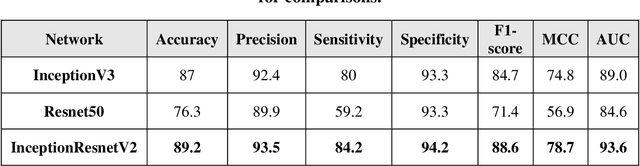
Prostate cancer is the most dangerous cancer diagnosed in men worldwide. Prostate diagnosis has been affected by many factors, such as lesion complexity, observer visibility, and variability. Many techniques based on Magnetic Resonance Imaging (MRI) have been used for prostate cancer identification and classification in the last few decades. Developing these techniques is crucial and has a great medical effect because they improve the treatment benefits and the chance of patients' survival. A new technique that depends on MRI has been proposed to improve the diagnosis. This technique consists of two stages. First, the MRI images have been preprocessed to make the medical image more suitable for the detection step. Second, prostate cancer identification has been performed based on a pre-trained deep learning model, InceptionResNetV2, that has many advantages and achieves effective results. In this paper, the InceptionResNetV2 deep learning model used for this purpose has average accuracy equals to 89.20%, and the area under the curve (AUC) equals to 93.6%. The experimental results of this proposed new deep learning technique represent promising and effective results compared to other previous techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge