"cancer detection": models, code, and papers
Deep Learning Hyperparameter Optimization for Breast Mass Detection in Mammograms
Jul 22, 2022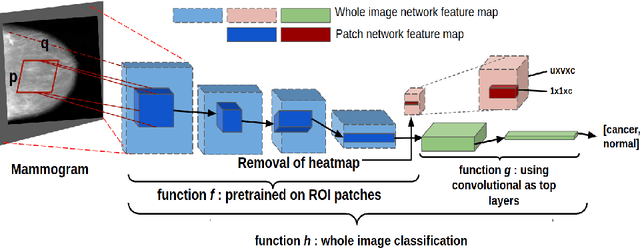
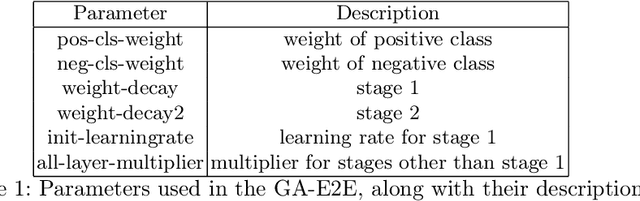
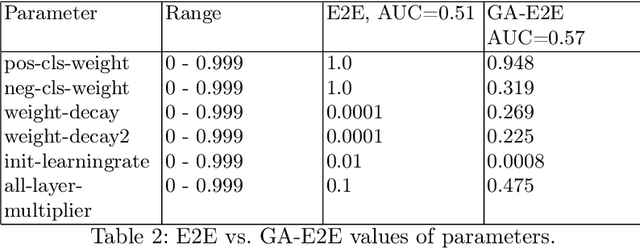
Accurate breast cancer diagnosis through mammography has the potential to save millions of lives around the world. Deep learning (DL) methods have shown to be very effective for mass detection in mammograms. Additional improvements of current DL models will further improve the effectiveness of these methods. A critical issue in this context is how to pick the right hyperparameters for DL models. In this paper, we present GA-E2E, a new approach for tuning the hyperparameters of DL models for brest cancer detection using Genetic Algorithms (GAs). Our findings reveal that differences in parameter values can considerably alter the area under the curve (AUC), which is used to determine a classifier's performance.
A Method to Facilitate Cancer Detection and Type Classification from Gene Expression Data using a Deep Autoencoder and Neural Network
Dec 20, 2018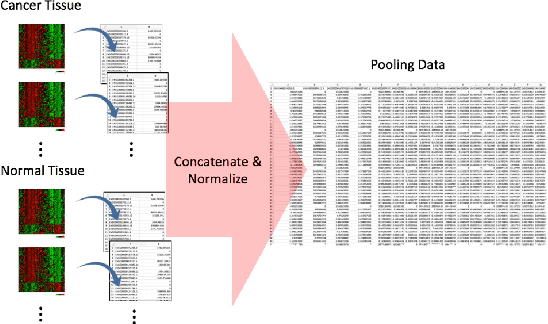
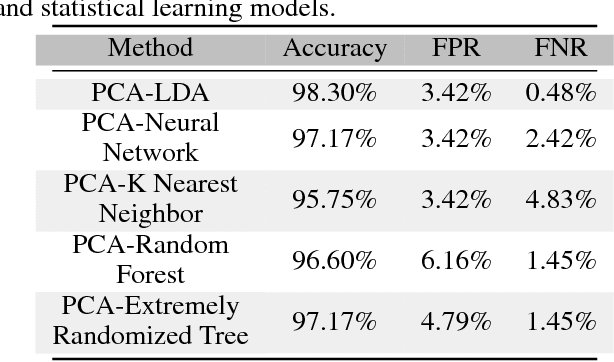
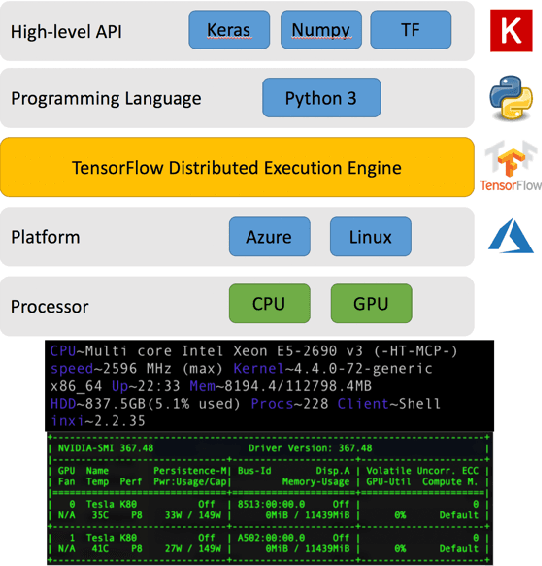
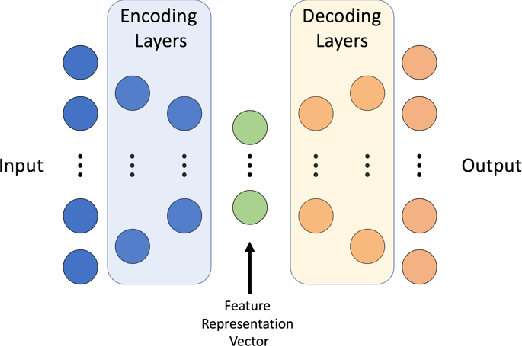
With the increased affordability and availability of whole-genome sequencing, large-scale and high-throughput gene expression is widely used to characterize diseases, including cancers. However, establishing specificity in cancer diagnosis using gene expression data continues to pose challenges due to the high dimensionality and complexity of the data. Here we present models of deep learning (DL) and apply them to gene expression data for the diagnosis and categorization of cancer. In this study, we have developed two DL models using messenger ribonucleic acid (mRNA) datasets available from the Genomic Data Commons repository. Our models achieved 98% accuracy in cancer detection, with false negative and false positive rates below 1.7%. In our results, we demonstrated that 18 out of 32 cancer-typing classifications achieved more than 90% accuracy. Due to the limitation of a small sample size (less than 50 observations), certain cancers could not achieve a higher accuracy in typing classification, but still achieved high accuracy for the cancer detection task. To validate our models, we compared them with traditional statistical models. The main advantage of our models over traditional cancer detection is the ability to use data from various cancer types to automatically form features to enhance the detection and diagnosis of a specific cancer type.
Genetic Analysis of Prostate Cancer with Computer Science Methods
Mar 29, 2023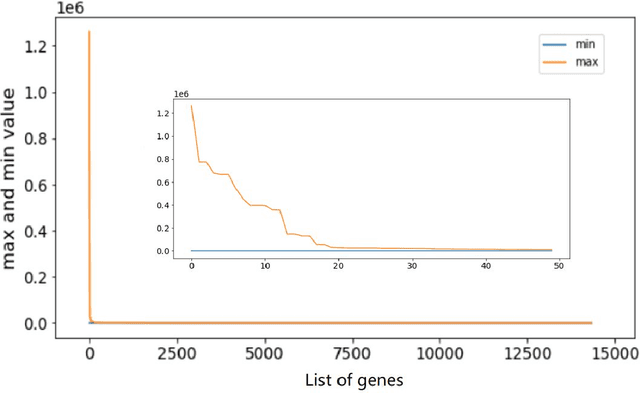
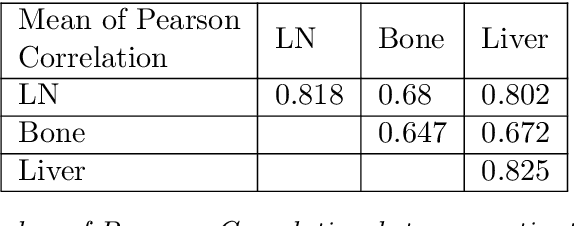
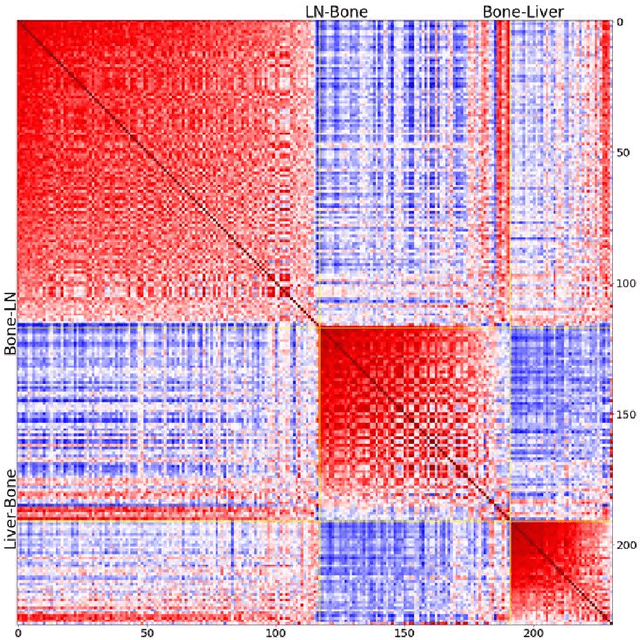
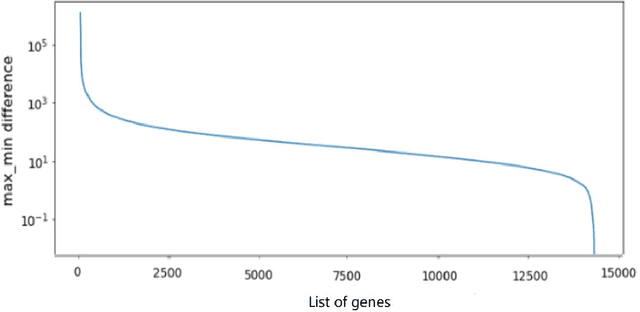
Metastatic prostate cancer is one of the most common cancers in men. In the advanced stages of prostate cancer, tumours can metastasise to other tissues in the body, which is fatal. In this thesis, we performed a genetic analysis of prostate cancer tumours at different metastatic sites using data science, machine learning and topological network analysis methods. We presented a general procedure for pre-processing gene expression datasets and pre-filtering significant genes by analytical methods. We then used machine learning models for further key gene filtering and secondary site tumour classification. Finally, we performed gene co-expression network analysis and community detection on samples from different prostate cancer secondary site types. In this work, 13 of the 14,379 genes were selected as the most metastatic prostate cancer related genes, achieving approximately 92% accuracy under cross-validation. In addition, we provide preliminary insights into the co-expression patterns of genes in gene co-expression networks. Project code is available at https://github.com/zcablii/Master_cancer_project.
Enhancing Early Lung Cancer Detection on Chest Radiographs with AI-assistance: A Multi-Reader Study
Aug 31, 2022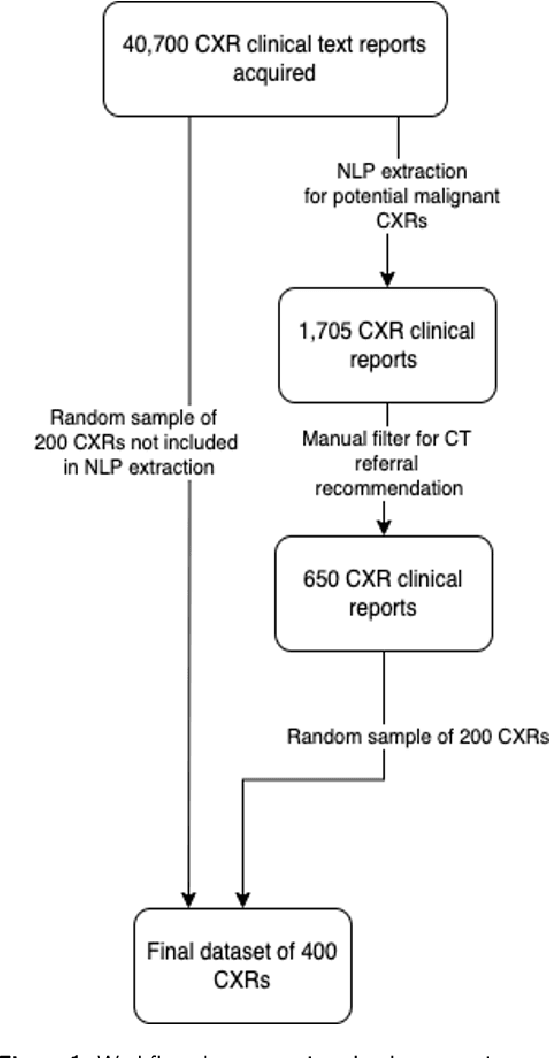
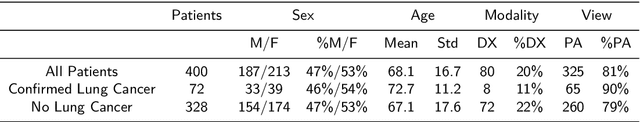
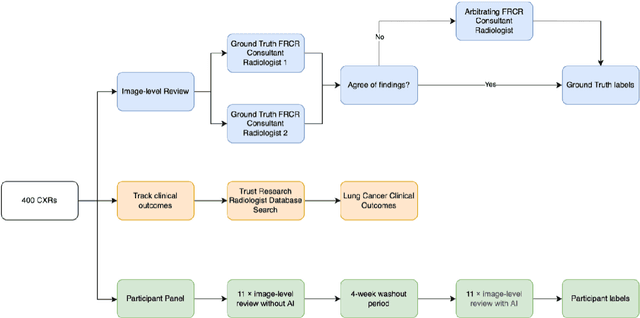
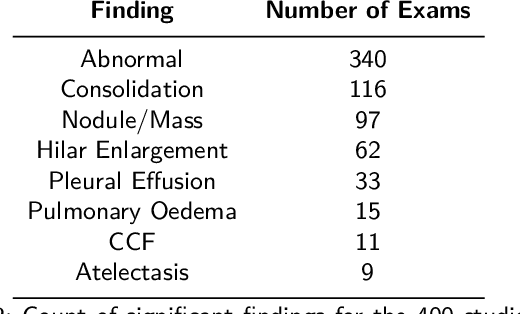
Objectives: The present study evaluated the impact of a commercially available explainable AI algorithm in augmenting the ability of clinicians to identify lung cancer on chest X-rays (CXR). Design: This retrospective study evaluated the performance of 11 clinicians for detecting lung cancer from chest radiographs, with and without assistance from a commercially available AI algorithm (red dot, Behold.ai) that predicts suspected lung cancer from CXRs. Clinician performance was evaluated against clinically confirmed diagnoses. Setting: The study analysed anonymised patient data from an NHS hospital; the dataset consisted of 400 chest radiographs from adult patients (18 years and above) who had a CXR performed in 2020, with corresponding clinical text reports. Participants: A panel of readers consisting of 11 clinicians (consultant radiologists, radiologist trainees and reporting radiographers) participated in this study. Main outcome measures: Overall accuracy, sensitivity, specificity and precision for detecting lung cancer on CXRs by clinicians, with and without AI input. Agreement rates between clinicians and performance standard deviation were also evaluated, with and without AI input. Results: The use of the AI algorithm by clinicians led to an improved overall performance for lung tumour detection, achieving an overall increase of 17.4% of lung cancers being identified on CXRs which would have otherwise been missed, an overall increase in detection of smaller tumours, a 24% and 13% increased detection of stage 1 and stage 2 lung cancers respectively, and standardisation of clinician performance. Conclusions: This study showed great promise in the clinical utility of AI algorithms in improving early lung cancer diagnosis and promoting health equity through overall improvement in reader performances, without impacting downstream imaging resources.
RADIFUSION: A multi-radiomics deep learning based breast cancer risk prediction model using sequential mammographic images with image attention and bilateral asymmetry refinement
Apr 01, 2023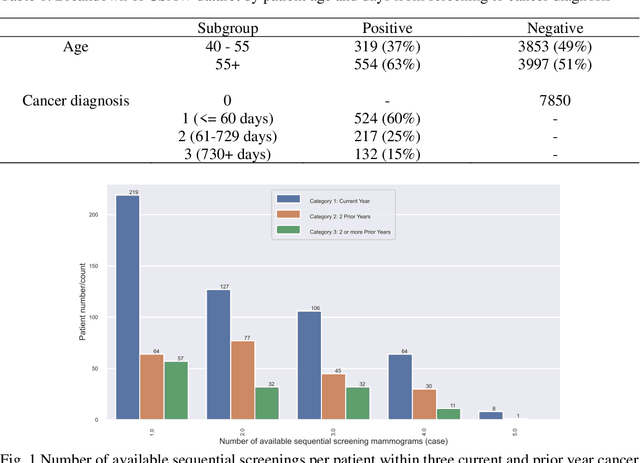
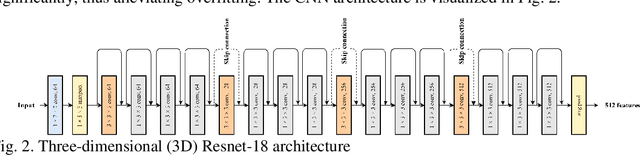
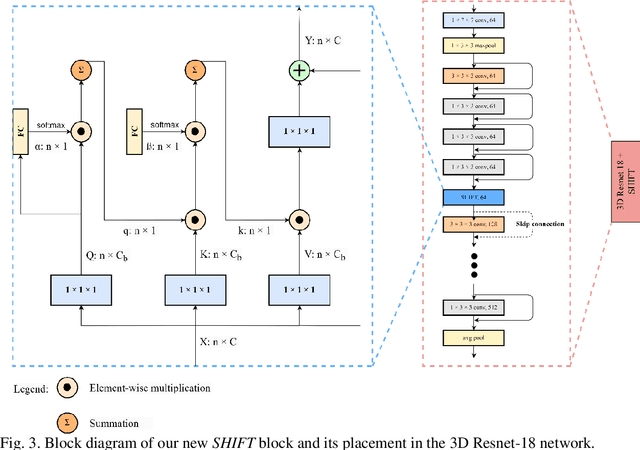

Breast cancer is a significant public health concern and early detection is critical for triaging high risk patients. Sequential screening mammograms can provide important spatiotemporal information about changes in breast tissue over time. In this study, we propose a deep learning architecture called RADIFUSION that utilizes sequential mammograms and incorporates a linear image attention mechanism, radiomic features, a new gating mechanism to combine different mammographic views, and bilateral asymmetry-based finetuning for breast cancer risk assessment. We evaluate our model on a screening dataset called Cohort of Screen-Aged Women (CSAW) dataset. Based on results obtained on the independent testing set consisting of 1,749 women, our approach achieved superior performance compared to other state-of-the-art models with area under the receiver operating characteristic curves (AUCs) of 0.905, 0.872 and 0.866 in the three respective metrics of 1-year AUC, 2-year AUC and > 2-year AUC. Our study highlights the importance of incorporating various deep learning mechanisms, such as image attention, radiomic features, gating mechanism, and bilateral asymmetry-based fine-tuning, to improve the accuracy of breast cancer risk assessment. We also demonstrate that our model's performance was enhanced by leveraging spatiotemporal information from sequential mammograms. Our findings suggest that RADIFUSION can provide clinicians with a powerful tool for breast cancer risk assessment.
Fluorescence molecular optomic signatures improve identification of tumors in head and neck specimens
Aug 29, 2022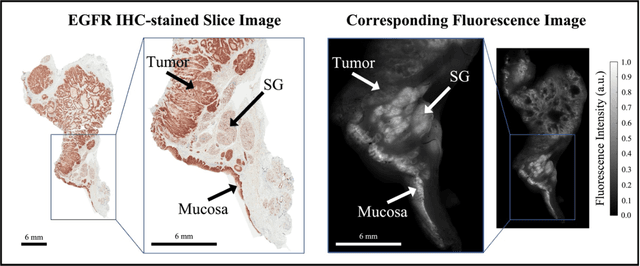
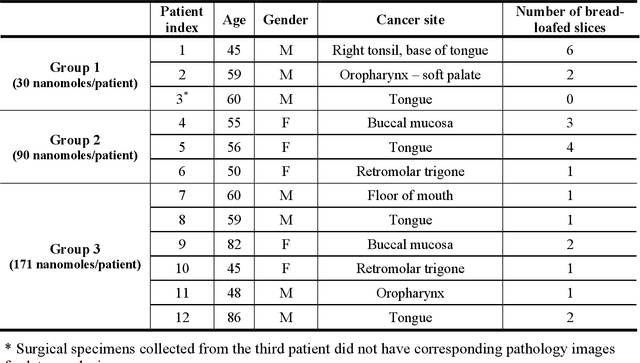
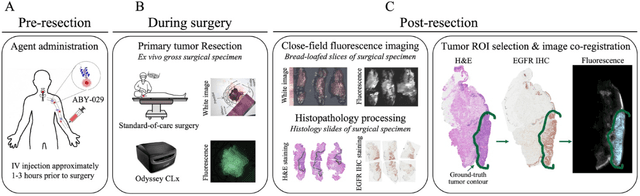
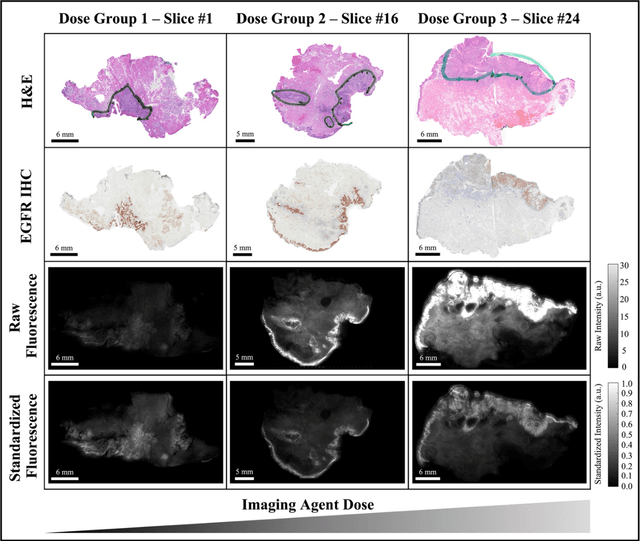
In this study, a radiomics approach was extended to optical fluorescence molecular imaging data for tissue classification, termed 'optomics'. Fluorescence molecular imaging is emerging for precise surgical guidance during head and neck squamous cell carcinoma (HNSCC) resection. However, the tumor-to-normal tissue contrast is confounded by intrinsic physiological limitations of heterogeneous expression of the target molecule, epidermal growth factor receptor (EGFR). Optomics seek to improve tumor identification by probing textural pattern differences in EGFR expression conveyed by fluorescence. A total of 1,472 standardized optomic features were extracted from fluorescence image samples. A supervised machine learning pipeline involving a support vector machine classifier was trained with 25 top-ranked features selected by minimum redundancy maximum relevance criterion. Model predictive performance was compared to fluorescence intensity thresholding method by classifying testing set image patches of resected tissue with histologically confirmed malignancy status. The optomics approach provided consistent improvement in prediction accuracy on all test set samples, irrespective of dose, compared to fluorescence intensity thresholding method (mean accuracies of 89% vs. 81%; P = 0.0072). The improved performance demonstrates that extending the radiomics approach to fluorescence molecular imaging data offers a promising image analysis technique for cancer detection in fluorescence-guided surgery.
Circular Antenna Array Design for Breast Cancer Detection
Jan 15, 2018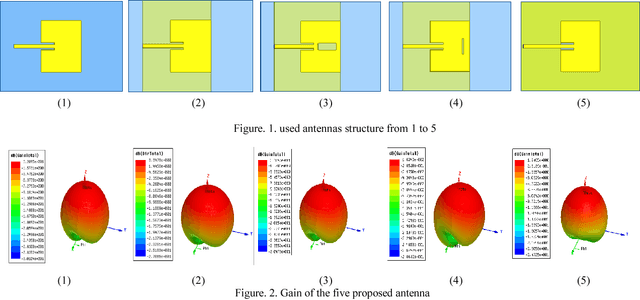
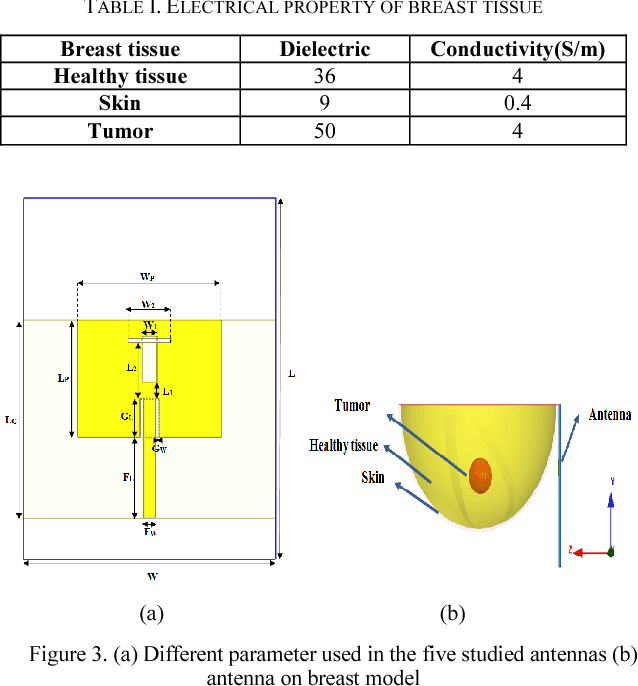
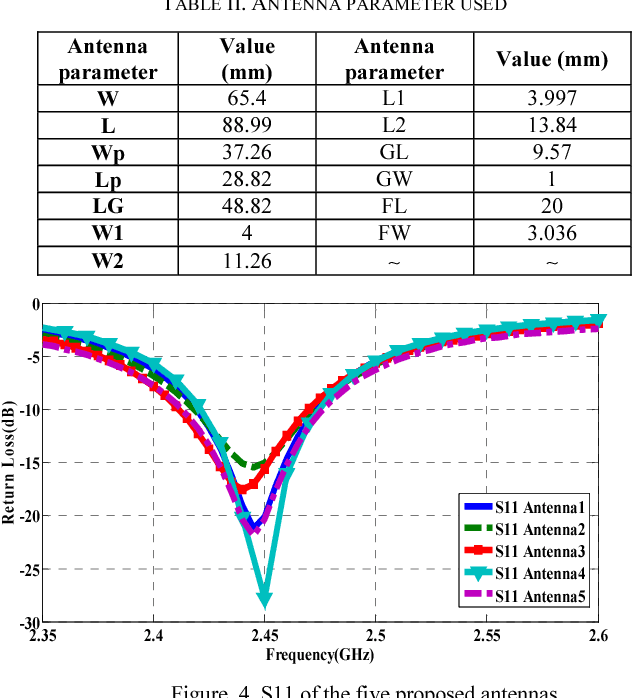
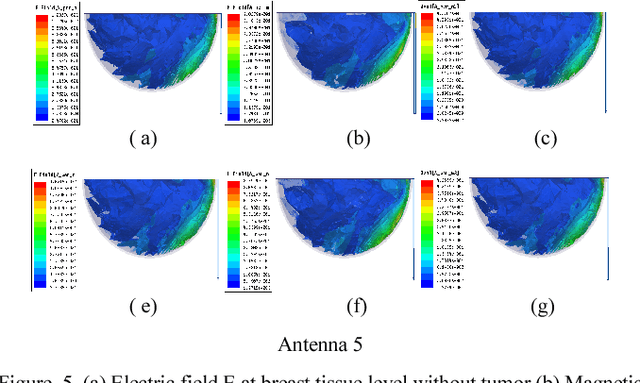
Microwave imaging for breast cancer detection is based on the contrast in the electrical properties of healthy fatty breast tissues. This paper presents an industrial, scientific and medical (ISM) bands comparative study of five microstrip patch antennas for microwave imaging at a frequency of 2.45 GHz. The choice of one antenna is made for an antenna array composed of 8 antennas for a microwave breast imaging system. Each antenna element is arranged in a circular configuration so that it can be directly faced to the breast phantom for better tumor detection. This choice is made by putting each antenna alone on the Breast skin to study the electric field, magnetic fields and current density in the healthy tissue of the breast phantom designed and simulated in Ansoft High Frequency Simulation Software (HFSS).
Meta-information-aware Dual-path Transformer for Differential Diagnosis of Multi-type Pancreatic Lesions in Multi-phase CT
Mar 02, 2023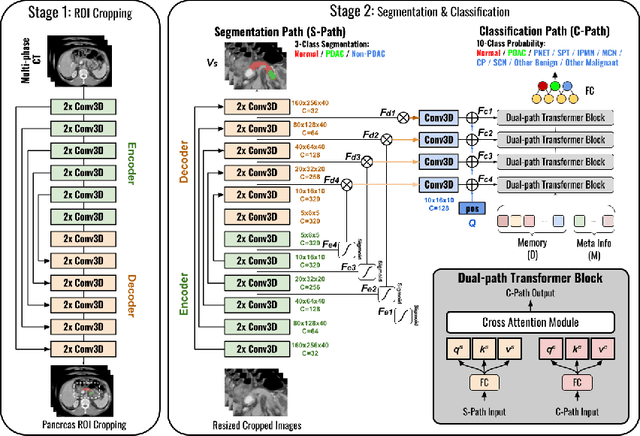
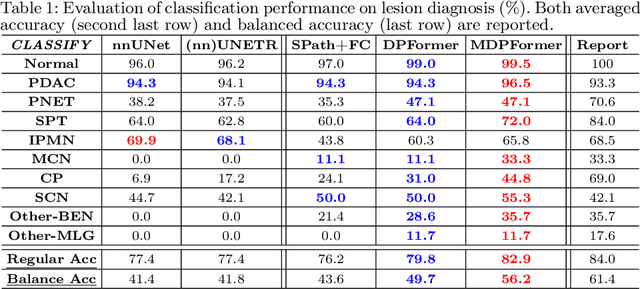
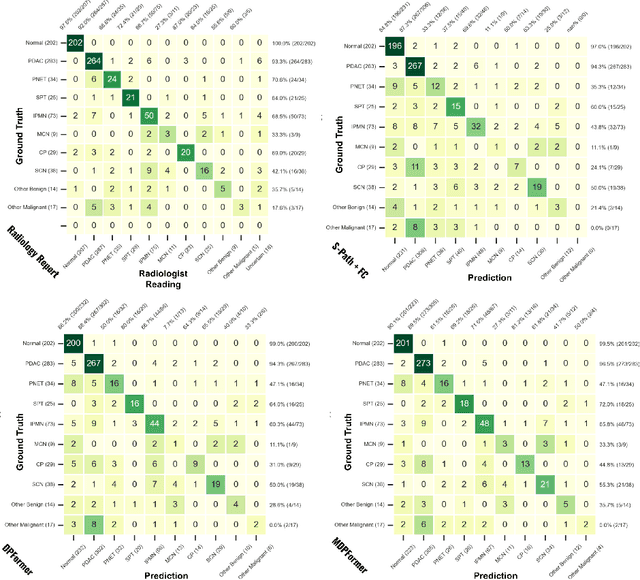
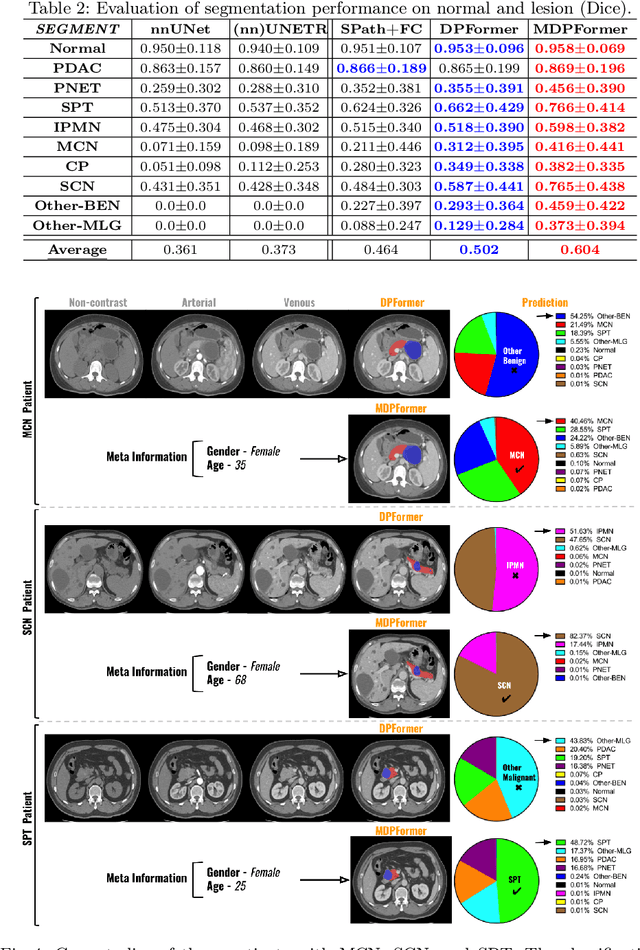
Pancreatic cancer is one of the leading causes of cancer-related death. Accurate detection, segmentation, and differential diagnosis of the full taxonomy of pancreatic lesions, i.e., normal, seven major types of lesions, and other lesions, is critical to aid the clinical decision-making of patient management and treatment. However, existing works focus on segmentation and classification for very specific lesion types (PDAC) or groups. Moreover, none of the previous work considers using lesion prevalence-related non-imaging patient information to assist the differential diagnosis. To this end, we develop a meta-information-aware dual-path transformer and exploit the feasibility of classification and segmentation of the full taxonomy of pancreatic lesions. Specifically, the proposed method consists of a CNN-based segmentation path (S-path) and a transformer-based classification path (C-path). The S-path focuses on initial feature extraction by semantic segmentation using a UNet-based network. The C-path utilizes both the extracted features and meta-information for patient-level classification based on stacks of dual-path transformer blocks that enhance the modeling of global contextual information. A large-scale multi-phase CT dataset of 3,096 patients with pathology-confirmed pancreatic lesion class labels, voxel-wise manual annotations of lesions from radiologists, and patient meta-information, was collected for training and evaluations. Our results show that our method can enable accurate classification and segmentation of the full taxonomy of pancreatic lesions, approaching the accuracy of the radiologist's report and significantly outperforming previous baselines. Results also show that adding the common meta-information, i.e., gender and age, can boost the model's performance, thus demonstrating the importance of meta-information for aiding pancreatic disease diagnosis.
EMT-NET: Efficient multitask network for computer-aided diagnosis of breast cancer
Jan 13, 2022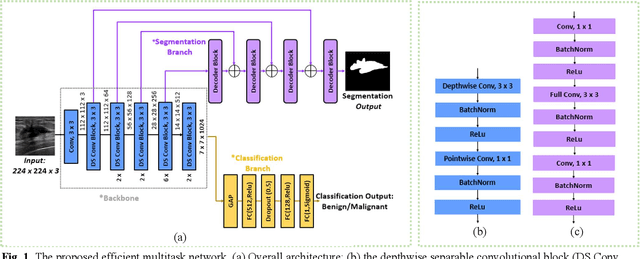
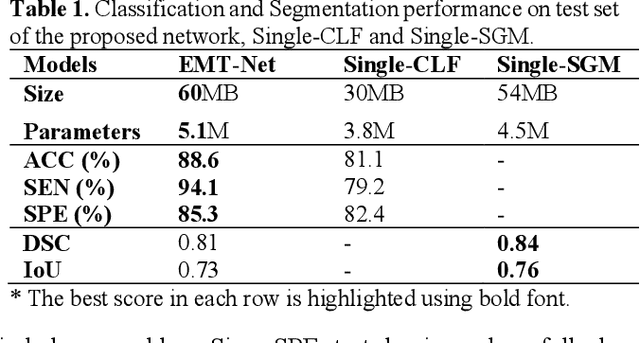
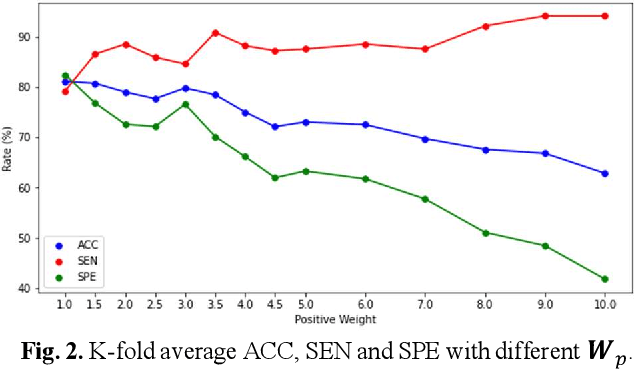
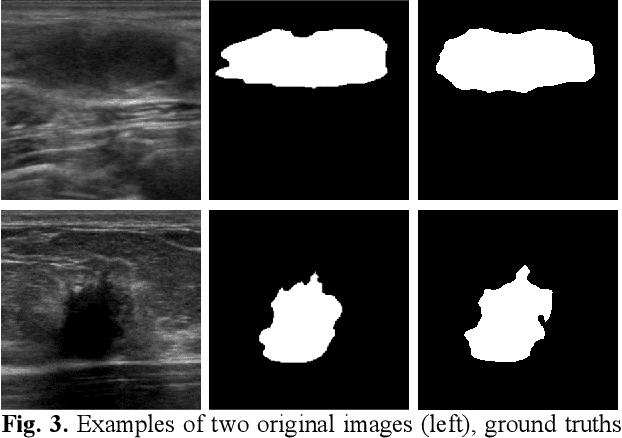
Deep learning-based computer-aided diagnosis has achieved unprecedented performance in breast cancer detection. However, most approaches are computationally intensive, which impedes their broader dissemination in real-world applications. In this work, we propose an efficient and light-weighted multitask learning architecture to classify and segment breast tumors simultaneously. We incorporate a segmentation task into a tumor classification network, which makes the backbone network learn representations focused on tumor regions. Moreover, we propose a new numerically stable loss function that easily controls the balance between the sensitivity and specificity of cancer detection. The proposed approach is evaluated using a breast ultrasound dataset with 1,511 images. The accuracy, sensitivity, and specificity of tumor classification is 88.6%, 94.1%, and 85.3%, respectively. We validate the model using a virtual mobile device, and the average inference time is 0.35 seconds per image.
Efficient High-Resolution Deep Learning: A Survey
Jul 26, 2022
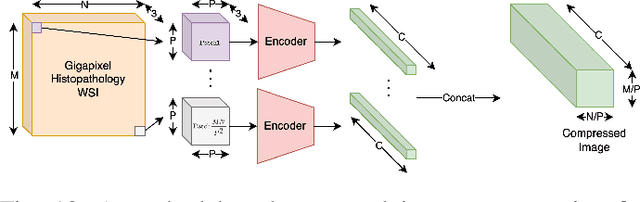

Cameras in modern devices such as smartphones, satellites and medical equipment are capable of capturing very high resolution images and videos. Such high-resolution data often need to be processed by deep learning models for cancer detection, automated road navigation, weather prediction, surveillance, optimizing agricultural processes and many other applications. Using high-resolution images and videos as direct inputs for deep learning models creates many challenges due to their high number of parameters, computation cost, inference latency and GPU memory consumption. Simple approaches such as resizing the images to a lower resolution are common in the literature, however, they typically significantly decrease accuracy. Several works in the literature propose better alternatives in order to deal with the challenges of high-resolution data and improve accuracy and speed while complying with hardware limitations and time restrictions. This survey describes such efficient high-resolution deep learning methods, summarizes real-world applications of high-resolution deep learning, and provides comprehensive information about available high-resolution datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge