"Information Extraction": models, code, and papers
Probing Representations for Document-level Event Extraction
Oct 23, 2023The probing classifiers framework has been employed for interpreting deep neural network models for a variety of natural language processing (NLP) applications. Studies, however, have largely focused on sentencelevel NLP tasks. This work is the first to apply the probing paradigm to representations learned for document-level information extraction (IE). We designed eight embedding probes to analyze surface, semantic, and event-understanding capabilities relevant to document-level event extraction. We apply them to the representations acquired by learning models from three different LLM-based document-level IE approaches on a standard dataset. We found that trained encoders from these models yield embeddings that can modestly improve argument detections and labeling but only slightly enhance event-level tasks, albeit trade-offs in information helpful for coherence and event-type prediction. We further found that encoder models struggle with document length and cross-sentence discourse.
A Review of Reinforcement Learning for Natural Language Processing, and Applications in Healthcare
Oct 23, 2023Reinforcement learning (RL) has emerged as a powerful approach for tackling complex medical decision-making problems such as treatment planning, personalized medicine, and optimizing the scheduling of surgeries and appointments. It has gained significant attention in the field of Natural Language Processing (NLP) due to its ability to learn optimal strategies for tasks such as dialogue systems, machine translation, and question-answering. This paper presents a review of the RL techniques in NLP, highlighting key advancements, challenges, and applications in healthcare. The review begins by visualizing a roadmap of machine learning and its applications in healthcare. And then it explores the integration of RL with NLP tasks. We examined dialogue systems where RL enables the learning of conversational strategies, RL-based machine translation models, question-answering systems, text summarization, and information extraction. Additionally, ethical considerations and biases in RL-NLP systems are addressed.
Exploring OCR Capabilities of GPT-4V(ision) : A Quantitative and In-depth Evaluation
Oct 29, 2023This paper presents a comprehensive evaluation of the Optical Character Recognition (OCR) capabilities of the recently released GPT-4V(ision), a Large Multimodal Model (LMM). We assess the model's performance across a range of OCR tasks, including scene text recognition, handwritten text recognition, handwritten mathematical expression recognition, table structure recognition, and information extraction from visually-rich document. The evaluation reveals that GPT-4V performs well in recognizing and understanding Latin contents, but struggles with multilingual scenarios and complex tasks. Specifically, it showed limitations when dealing with non-Latin languages and complex tasks such as handwriting mathematical expression recognition, table structure recognition, and end-to-end semantic entity recognition and pair extraction from document image. Based on these observations, we affirm the necessity and continued research value of specialized OCR models. In general, despite its versatility in handling diverse OCR tasks, GPT-4V does not outperform existing state-of-the-art OCR models. How to fully utilize pre-trained general-purpose LMMs such as GPT-4V for OCR downstream tasks remains an open problem. The study offers a critical reference for future research in OCR with LMMs. Evaluation pipeline and results are available at https://github.com/SCUT-DLVCLab/GPT-4V_OCR.
CAW-coref: Conjunction-Aware Word-level Coreference Resolution
Oct 19, 2023State-of-the-art coreference resolutions systems depend on multiple LLM calls per document and are thus prohibitively expensive for many use cases (e.g., information extraction with large corpora). The leading word-level coreference system (WL-coref) attains 96.6% of these SOTA systems' performance while being much more efficient. In this work, we identify a routine yet important failure case of WL-coref: dealing with conjoined mentions such as 'Tom and Mary'. We offer a simple yet effective solution that improves the performance on the OntoNotes test set by 0.9% F1, shrinking the gap between efficient word-level coreference resolution and expensive SOTA approaches by 34.6%. Our Conjunction-Aware Word-level coreference model (CAW-coref) and code is available at https://github.com/KarelDO/wl-coref.
Hierarchy Builder: Organizing Textual Spans into a Hierarchy to Facilitate Navigation
Sep 18, 2023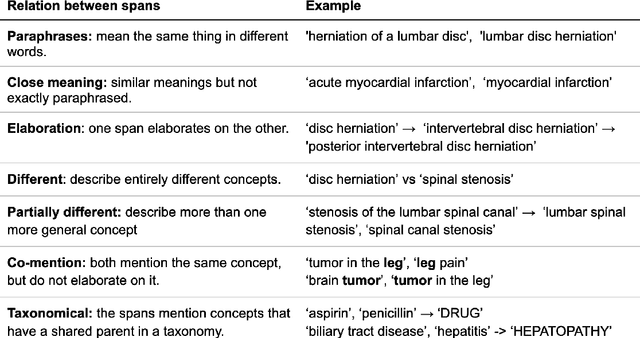
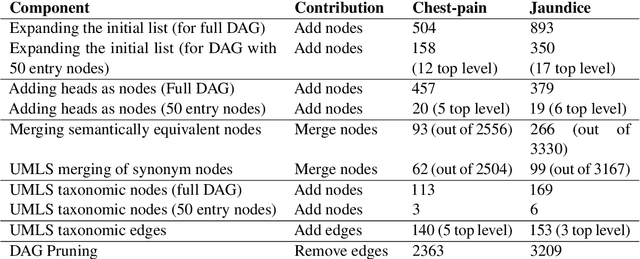
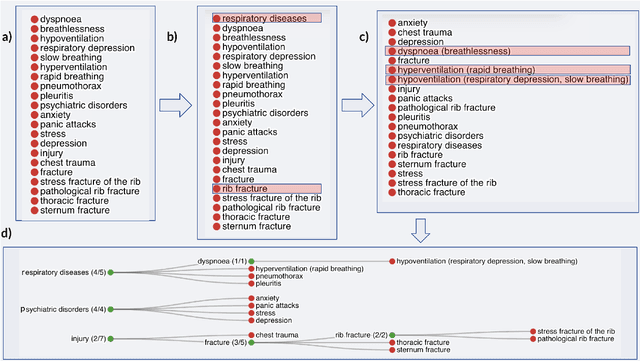
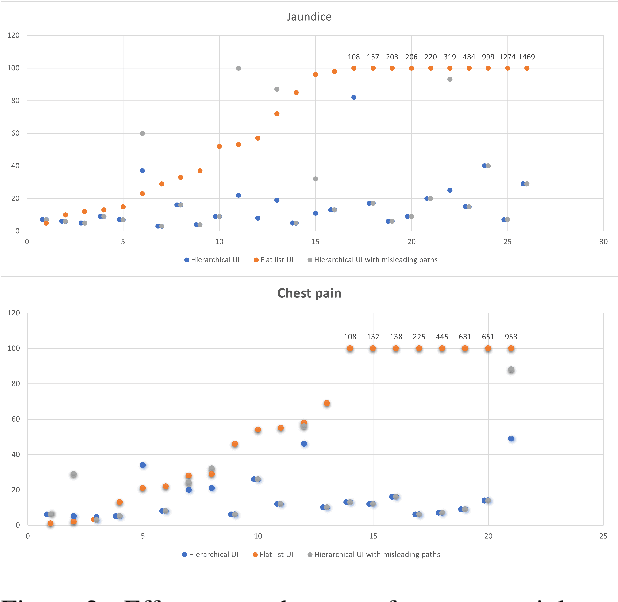
Information extraction systems often produce hundreds to thousands of strings on a specific topic. We present a method that facilitates better consumption of these strings, in an exploratory setting in which a user wants to both get a broad overview of what's available, and a chance to dive deeper on some aspects. The system works by grouping similar items together and arranging the remaining items into a hierarchical navigable DAG structure. We apply the method to medical information extraction.
* 9 pages including citations; Presented at the ACL 2023 DEMO track, pages 282-290
Rebuild City Buildings from Off-Nadir Aerial Images with Offset-Building Model (OBM)
Nov 03, 2023Accurate measurement of the offset from roof-to-footprint in very-high-resolution remote sensing imagery is crucial for urban information extraction tasks. With the help of deep learning, existing methods typically rely on two-stage CNN models to extract regions of interest on building feature maps. At the first stage, a Region Proposal Network (RPN) is applied to extract thousands of ROIs (Region of Interests) which will post-imported into a Region-based Convolutional Neural Networks (RCNN) to extract wanted information. However, because of inflexible RPN, these methods often lack effective user interaction, encounter difficulties in instance correspondence, and struggle to keep up with the advancements in general artificial intelligence. This paper introduces an interactive Transformer model combined with a prompt encoder to precisely extract building segmentation as well as the offset vectors from roofs to footprints. In our model, a powerful module, namely ROAM, was tailored for common problems in predicting roof-to-footprint offsets. We tested our model's feasibility on the publicly available BONAI dataset, achieving a significant reduction in Prompt-Instance-Level offset errors ranging from 14.6% to 16.3%. Additionally, we developed a Distance-NMS algorithm tailored for large-scale building offsets, significantly enhancing the accuracy of predicted building offset angles and lengths in a straightforward and efficient manner. To further validate the model's robustness, we created a new test set using 0.5m remote sensing imagery from Huizhou, China, for inference testing. Our code, training methods, and the updated dataset will be accessable at https://github.com/likaiucas.
Calibrated Seq2seq Models for Efficient and Generalizable Ultra-fine Entity Typing
Nov 01, 2023Ultra-fine entity typing plays a crucial role in information extraction by predicting fine-grained semantic types for entity mentions in text. However, this task poses significant challenges due to the massive number of entity types in the output space. The current state-of-the-art approaches, based on standard multi-label classifiers or cross-encoder models, suffer from poor generalization performance or inefficient inference. In this paper, we present CASENT, a seq2seq model designed for ultra-fine entity typing that predicts ultra-fine types with calibrated confidence scores. Our model takes an entity mention as input and employs constrained beam search to generate multiple types autoregressively. The raw sequence probabilities associated with the predicted types are then transformed into confidence scores using a novel calibration method. We conduct extensive experiments on the UFET dataset which contains over 10k types. Our method outperforms the previous state-of-the-art in terms of F1 score and calibration error, while achieving an inference speedup of over 50 times. Additionally, we demonstrate the generalization capabilities of our model by evaluating it in zero-shot and few-shot settings on five specialized domain entity typing datasets that are unseen during training. Remarkably, our model outperforms large language models with 10 times more parameters in the zero-shot setting, and when fine-tuned on 50 examples, it significantly outperforms ChatGPT on all datasets. Our code, models and demo are available at https://github.com/yanlinf/CASENT.
Joint Entity and Relation Extraction with Span Pruning and Hypergraph Neural Networks
Oct 26, 2023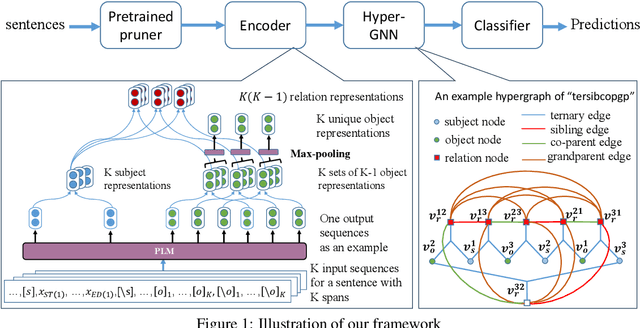
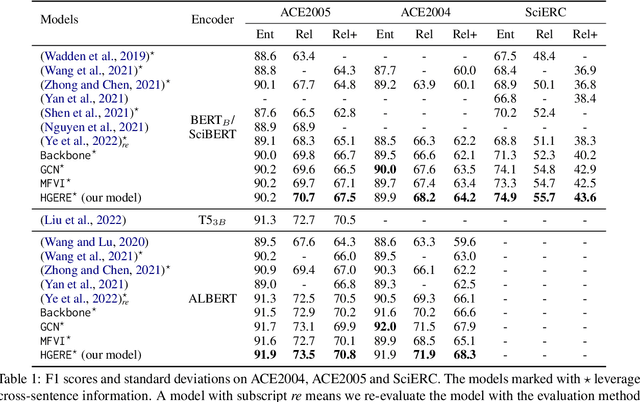
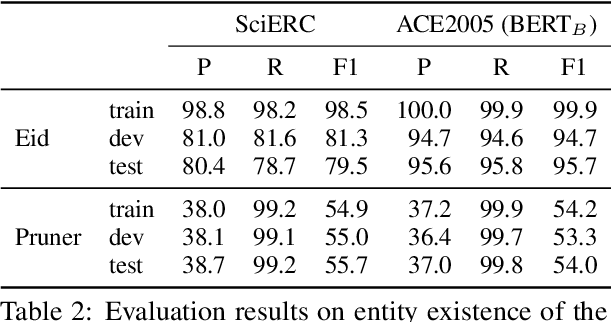
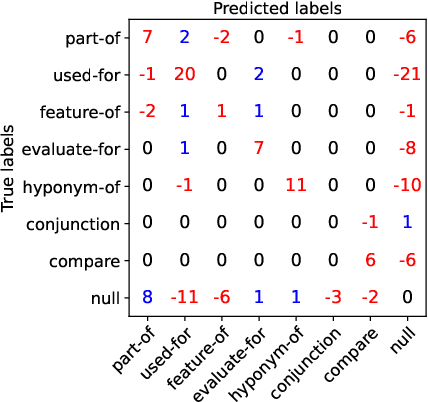
Entity and Relation Extraction (ERE) is an important task in information extraction. Recent marker-based pipeline models achieve state-of-the-art performance, but still suffer from the error propagation issue. Also, most of current ERE models do not take into account higher-order interactions between multiple entities and relations, while higher-order modeling could be beneficial.In this work, we propose HyperGraph neural network for ERE ($\hgnn{}$), which is built upon the PL-marker (a state-of-the-art marker-based pipleline model). To alleviate error propagation,we use a high-recall pruner mechanism to transfer the burden of entity identification and labeling from the NER module to the joint module of our model. For higher-order modeling, we build a hypergraph, where nodes are entities (provided by the span pruner) and relations thereof, and hyperedges encode interactions between two different relations or between a relation and its associated subject and object entities. We then run a hypergraph neural network for higher-order inference by applying message passing over the built hypergraph. Experiments on three widely used benchmarks (\acef{}, \ace{} and \scierc{}) for ERE task show significant improvements over the previous state-of-the-art PL-marker.
Vision-Enhanced Semantic Entity Recognition in Document Images via Visually-Asymmetric Consistency Learning
Oct 23, 2023Extracting meaningful entities belonging to predefined categories from Visually-rich Form-like Documents (VFDs) is a challenging task. Visual and layout features such as font, background, color, and bounding box location and size provide important cues for identifying entities of the same type. However, existing models commonly train a visual encoder with weak cross-modal supervision signals, resulting in a limited capacity to capture these non-textual features and suboptimal performance. In this paper, we propose a novel \textbf{V}isually-\textbf{A}symmetric co\textbf{N}sisten\textbf{C}y \textbf{L}earning (\textsc{Vancl}) approach that addresses the above limitation by enhancing the model's ability to capture fine-grained visual and layout features through the incorporation of color priors. Experimental results on benchmark datasets show that our approach substantially outperforms the strong LayoutLM series baseline, demonstrating the effectiveness of our approach. Additionally, we investigate the effects of different color schemes on our approach, providing insights for optimizing model performance. We believe our work will inspire future research on multimodal information extraction.
Augmented Kinesthetic Teaching: Enhancing Task Execution Efficiency through Intuitive Human Instructions
Dec 01, 2023In this paper, we present a complete and efficient implementation of a knowledge-sharing augmented kinesthetic teaching approach for efficient task execution in robotics. Our augmented kinesthetic teaching method integrates intuitive human feedback, including verbal, gesture, gaze, and physical guidance, to facilitate the extraction of multiple layers of task information including control type, attention direction, input and output type, action state change trigger, etc., enhancing the adaptability and autonomy of robots during task execution. We propose an efficient Programming by Demonstration (PbD) framework for users with limited technical experience to teach the robot in an intuitive manner. The proposed framework provides an interface for such users to teach customized tasks using high-level commands, with the goal of achieving a smoother teaching experience and task execution. This is demonstrated with the sample task of pouring water.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge