"Information Extraction": models, code, and papers
Wasserstein Nonnegative Tensor Factorization with Manifold Regularization
Jan 03, 2024Nonnegative tensor factorization (NTF) has become an important tool for feature extraction and part-based representation with preserved intrinsic structure information from nonnegative high-order data. However, the original NTF methods utilize Euclidean or Kullback-Leibler divergence as the loss function which treats each feature equally leading to the neglect of the side-information of features. To utilize correlation information of features and manifold information of samples, we introduce Wasserstein manifold nonnegative tensor factorization (WMNTF), which minimizes the Wasserstein distance between the distribution of input tensorial data and the distribution of reconstruction. Although some researches about Wasserstein distance have been proposed in nonnegative matrix factorization (NMF), they ignore the spatial structure information of higher-order data. We use Wasserstein distance (a.k.a Earth Mover's distance or Optimal Transport distance) as a metric and add a graph regularizer to a latent factor. Experimental results demonstrate the effectiveness of the proposed method compared with other NMF and NTF methods.
Passive Integrated Sensing and Communication Scheme based on RF Fingerprint Information Extraction for Cell-Free RAN
Nov 10, 2023This paper investigates how to achieve integrated sensing and communication (ISAC) based on a cell-free radio access network (CF-RAN) architecture with a minimum footprint of communication resources. We propose a new passive sensing scheme. The scheme is based on the radio frequency (RF) fingerprint learning of the RF radio unit (RRU) to build an RF fingerprint library of RRUs. The source RRU is identified by comparing the RF fingerprints carried by the signal at the receiver side. The receiver extracts the channel parameters from the signal and estimates the channel environment, thus locating the reflectors in the environment. The proposed scheme can effectively solve the problem of interference between signals in the same time-frequency domain but in different spatial domains when multiple RRUs jointly serve users in CF-RAN architecture. Simulation results show that the proposed passive ISAC scheme can effectively detect reflector location information in the environment without degrading the communication performance.
FSUIE: A Novel Fuzzy Span Mechanism for Universal Information Extraction
Jun 19, 2023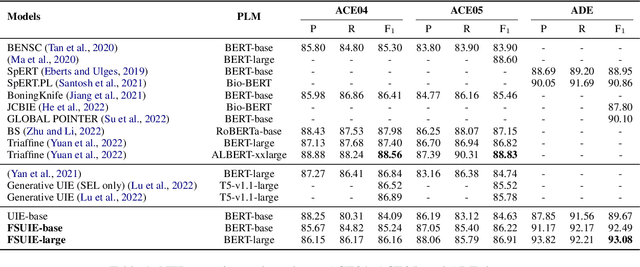
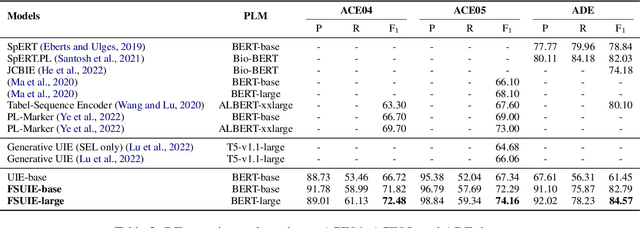
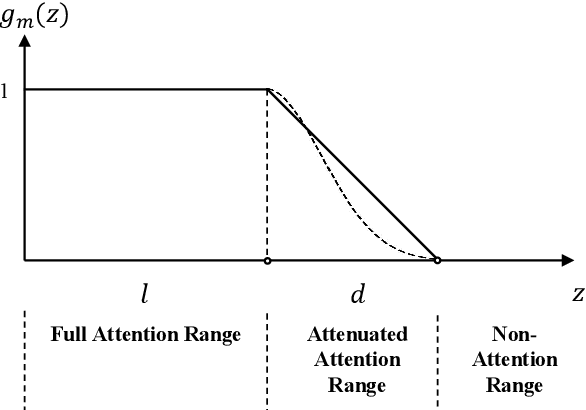
Universal Information Extraction (UIE) has been introduced as a unified framework for various Information Extraction (IE) tasks and has achieved widespread success. Despite this, UIE models have limitations. For example, they rely heavily on span boundaries in the data during training, which does not reflect the reality of span annotation challenges. Slight adjustments to positions can also meet requirements. Additionally, UIE models lack attention to the limited span length feature in IE. To address these deficiencies, we propose the Fuzzy Span Universal Information Extraction (FSUIE) framework. Specifically, our contribution consists of two concepts: fuzzy span loss and fuzzy span attention. Our experimental results on a series of main IE tasks show significant improvement compared to the baseline, especially in terms of fast convergence and strong performance with small amounts of data and training epochs. These results demonstrate the effectiveness and generalization of FSUIE in different tasks, settings, and scenarios.
AviationGPT: A Large Language Model for the Aviation Domain
Nov 29, 2023The advent of ChatGPT and GPT-4 has captivated the world with large language models (LLMs), demonstrating exceptional performance in question-answering, summarization, and content generation. The aviation industry is characterized by an abundance of complex, unstructured text data, replete with technical jargon and specialized terminology. Moreover, labeled data for model building are scarce in this domain, resulting in low usage of aviation text data. The emergence of LLMs presents an opportunity to transform this situation, but there is a lack of LLMs specifically designed for the aviation domain. To address this gap, we propose AviationGPT, which is built on open-source LLaMA-2 and Mistral architectures and continuously trained on a wealth of carefully curated aviation datasets. Experimental results reveal that AviationGPT offers users multiple advantages, including the versatility to tackle diverse natural language processing (NLP) problems (e.g., question-answering, summarization, document writing, information extraction, report querying, data cleaning, and interactive data exploration). It also provides accurate and contextually relevant responses within the aviation domain and significantly improves performance (e.g., over a 40% performance gain in tested cases). With AviationGPT, the aviation industry is better equipped to address more complex research problems and enhance the efficiency and safety of National Airspace System (NAS) operations.
VKIE: The Application of Key Information Extraction on Video Text
Oct 18, 2023Extracting structured information from videos is critical for numerous downstream applications in the industry. In this paper, we define a significant task of extracting hierarchical key information from visual texts on videos. To fulfill this task, we decouples it into four subtasks and introduce two implementation solutions called PipVKIE and UniVKIE. PipVKIE sequentially completes the four subtasks in continuous stages, while UniVKIE is improved by unifying all the subtasks into one backbone. Both PipVKIE and UniVKIE leverage multimodal information from vision, text, and coordinates for feature representation. Extensive experiments on one well-defined dataset demonstrate that our solutions can achieve remarkable performance and efficient inference speed. The code and dataset will be publicly available.
Similar Document Template Matching Algorithm
Nov 21, 2023This study outlines a comprehensive methodology for verifying medical documents, integrating advanced techniques in template extraction, comparison, and fraud detection. It begins with template extraction using sophisticated region-of-interest (ROI) methods, incorporating contour analysis and edge identification. Pre-processing steps ensure template clarity through morphological operations and adaptive thresholding. The template comparison algorithm utilizes advanced feature matching with key points and descriptors, enhancing robustness through histogram-based analysis for accounting variations. Fraud detection involves the SSIM computation and OCR for textual information extraction. The SSIM quantifies structural similarity, aiding in potential match identification. OCR focuses on critical areas like patient details, provider information, and billing amounts. Extracted information is compared with a reference dataset, and confidence thresholding ensures reliable fraud detection. Adaptive parameters enhance system flexibility for dynamic adjustments to varying document layouts. This methodology provides a robust approach to medical document verification, addressing complexities in template extraction, comparison, fraud detection, and adaptability to diverse document structures.
PPN: Parallel Pointer-based Network for Key Information Extraction with Complex Layouts
Jul 20, 2023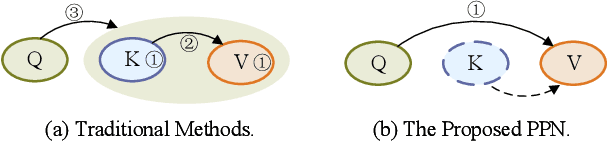
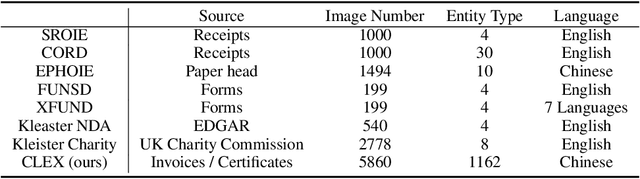
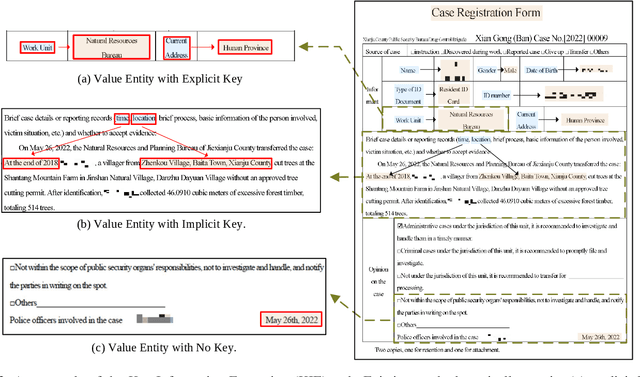
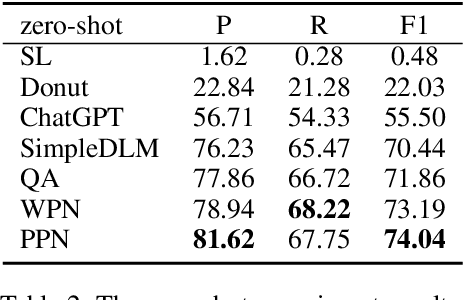
Key Information Extraction (KIE) is a challenging multimodal task that aims to extract structured value semantic entities from visually rich documents. Although significant progress has been made, there are still two major challenges that need to be addressed. Firstly, the layout of existing datasets is relatively fixed and limited in the number of semantic entity categories, creating a significant gap between these datasets and the complex real-world scenarios. Secondly, existing methods follow a two-stage pipeline strategy, which may lead to the error propagation problem. Additionally, they are difficult to apply in situations where unseen semantic entity categories emerge. To address the first challenge, we propose a new large-scale human-annotated dataset named Complex Layout form for key information EXtraction (CLEX), which consists of 5,860 images with 1,162 semantic entity categories. To solve the second challenge, we introduce Parallel Pointer-based Network (PPN), an end-to-end model that can be applied in zero-shot and few-shot scenarios. PPN leverages the implicit clues between semantic entities to assist extracting, and its parallel extraction mechanism allows it to extract multiple results simultaneously and efficiently. Experiments on the CLEX dataset demonstrate that PPN outperforms existing state-of-the-art methods while also offering a much faster inference speed.
Explanatory Argument Extraction of Correct Answers in Resident Medical Exams
Dec 01, 2023Developing the required technology to assist medical experts in their everyday activities is currently a hot topic in the Artificial Intelligence research field. Thus, a number of large language models (LLMs) and automated benchmarks have recently been proposed with the aim of facilitating information extraction in Evidence-Based Medicine (EBM) using natural language as a tool for mediating in human-AI interaction. The most representative benchmarks are limited to either multiple-choice or long-form answers and are available only in English. In order to address these shortcomings, in this paper we present a new dataset which, unlike previous work: (i) includes not only explanatory arguments for the correct answer, but also arguments to reason why the incorrect answers are not correct; (ii) the explanations are written originally by medical doctors to answer questions from the Spanish Residency Medical Exams. Furthermore, this new benchmark allows us to setup a novel extractive task which consists of identifying the explanation of the correct answer written by medical doctors. An additional benefit of our setting is that we can leverage the extractive QA paradigm to automatically evaluate performance of LLMs without resorting to costly manual evaluation by medical experts. Comprehensive experimentation with language models for Spanish shows that sometimes multilingual models fare better than monolingual ones, even outperforming models which have been adapted to the medical domain. Furthermore, results across the monolingual models are mixed, with supposedly smaller and inferior models performing competitively. In any case, the obtained results show that our novel dataset and approach can be an effective technique to help medical practitioners in identifying relevant evidence-based explanations for medical questions.
Improving embedding of graphs with missing data by soft manifolds
Nov 29, 2023Embedding graphs in continous spaces is a key factor in designing and developing algorithms for automatic information extraction to be applied in diverse tasks (e.g., learning, inferring, predicting). The reliability of graph embeddings directly depends on how much the geometry of the continuous space matches the graph structure. Manifolds are mathematical structure that can enable to incorporate in their topological spaces the graph characteristics, and in particular nodes distances. State-of-the-art of manifold-based graph embedding algorithms take advantage of the assumption that the projection on a tangential space of each point in the manifold (corresponding to a node in the graph) would locally resemble a Euclidean space. Although this condition helps in achieving efficient analytical solutions to the embedding problem, it does not represent an adequate set-up to work with modern real life graphs, that are characterized by weighted connections across nodes often computed over sparse datasets with missing records. In this work, we introduce a new class of manifold, named soft manifold, that can solve this situation. In particular, soft manifolds are mathematical structures with spherical symmetry where the tangent spaces to each point are hypocycloids whose shape is defined according to the velocity of information propagation across the data points. Using soft manifolds for graph embedding, we can provide continuous spaces to pursue any task in data analysis over complex datasets. Experimental results on reconstruction tasks on synthetic and real datasets show how the proposed approach enable more accurate and reliable characterization of graphs in continuous spaces with respect to the state-of-the-art.
Product Information Extraction using ChatGPT
Jun 23, 2023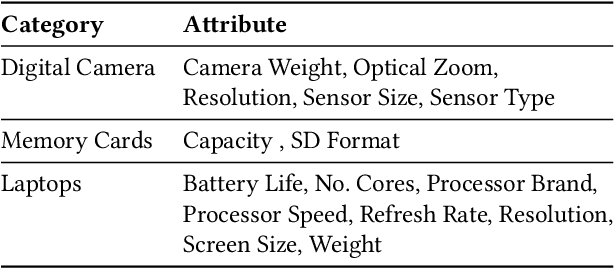
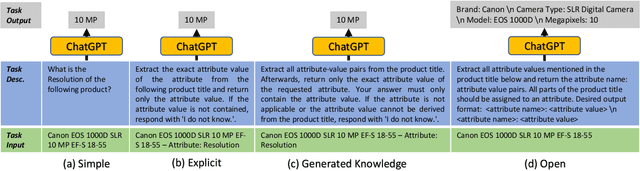
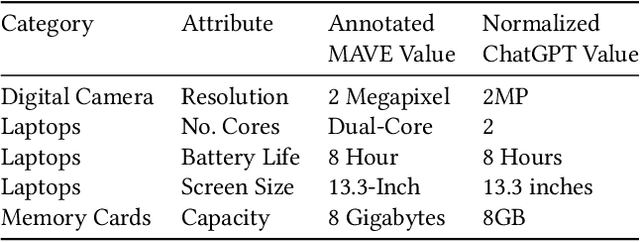
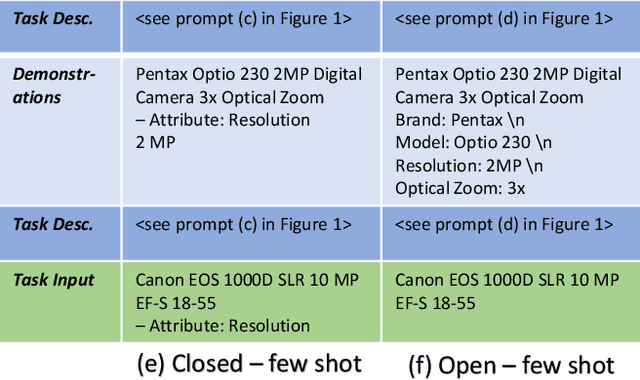
Structured product data in the form of attribute/value pairs is the foundation of many e-commerce applications such as faceted product search, product comparison, and product recommendation. Product offers often only contain textual descriptions of the product attributes in the form of titles or free text. Hence, extracting attribute/value pairs from textual product descriptions is an essential enabler for e-commerce applications. In order to excel, state-of-the-art product information extraction methods require large quantities of task-specific training data. The methods also struggle with generalizing to out-of-distribution attributes and attribute values that were not a part of the training data. Due to being pre-trained on huge amounts of text as well as due to emergent effects resulting from the model size, Large Language Models like ChatGPT have the potential to address both of these shortcomings. This paper explores the potential of ChatGPT for extracting attribute/value pairs from product descriptions. We experiment with different zero-shot and few-shot prompt designs. Our results show that ChatGPT achieves a performance similar to a pre-trained language model but requires much smaller amounts of training data and computation for fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge