"Information Extraction": models, code, and papers
EMOFM: Ensemble MLP mOdel with Feature-based Mixers for Click-Through Rate Prediction
Oct 15, 2023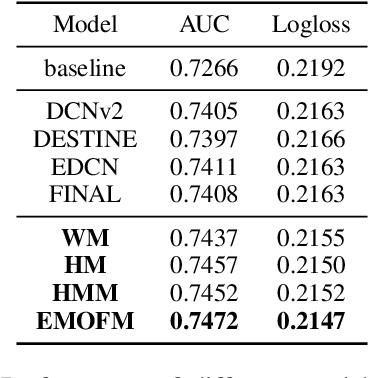
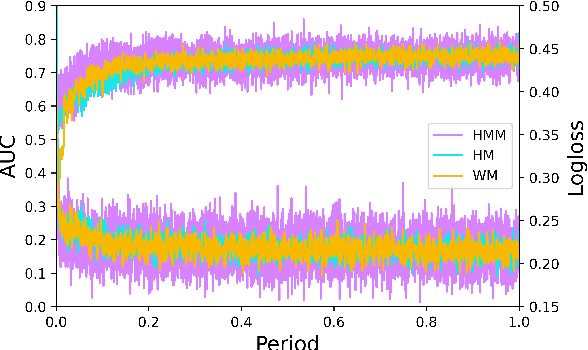
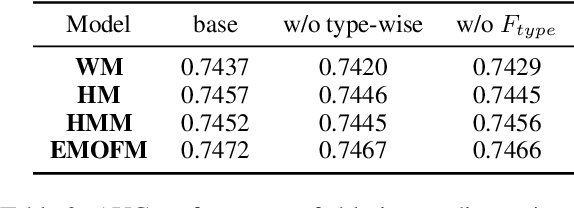
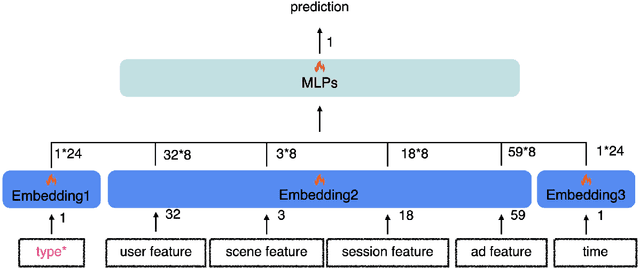
Track one of CTI competition is on click-through rate (CTR) prediction. The dataset contains millions of records and each field-wise feature in a record consists of hashed integers for privacy. For this task, the keys of network-based methods might be type-wise feature extraction and information fusion across different fields. Multi-layer perceptrons (MLPs) are able to extract field feature, but could not efficiently fuse features. Motivated by the natural fusion characteristic of cross attention and the efficiency of transformer-based structures, we propose simple plug-in mixers for field/type-wise feature fusion, and thus construct an field&type-wise ensemble model, namely EMOFM (Ensemble MLP mOdel with Feature-based Mixers). In the experiments, the proposed model is evaluated on the dataset, the optimization process is visualized and ablation studies are explored. It is shown that EMOFM outperforms compared baselines. In the end, we discuss on future work. WARNING: The comparison might not be fair enough since the proposed method is designed for this data in particular while compared methods are not. For example, EMOFM especially takes different types of interactions into consideration while others do not. Anyway, we do hope that the ideas inside our method could help other developers/learners/researchers/thinkers and so on.
HiNet: Novel Multi-Scenario & Multi-Task Learning with Hierarchical Information Extraction
Mar 14, 2023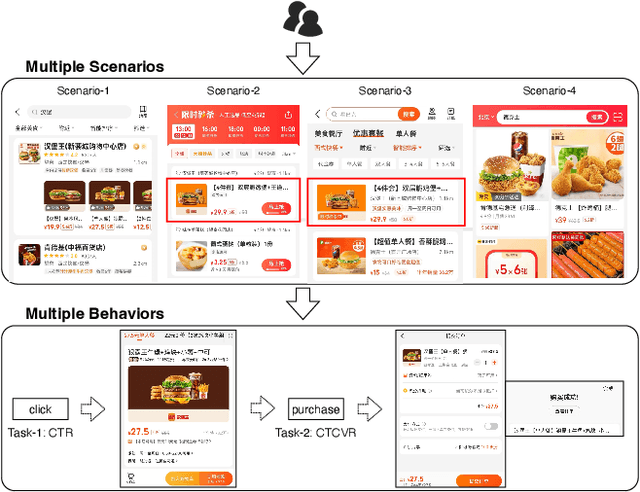
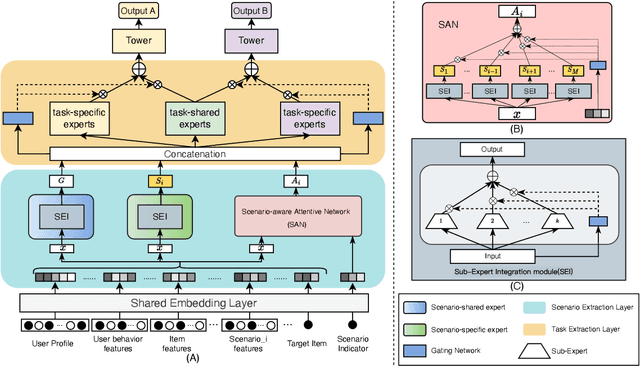
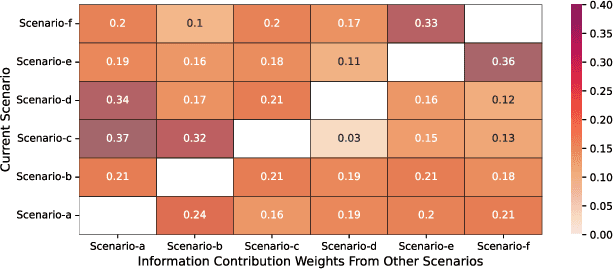
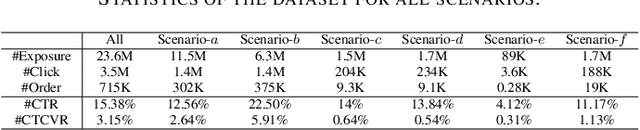
Multi-scenario & multi-task learning has been widely applied to many recommendation systems in industrial applications, wherein an effective and practical approach is to carry out multi-scenario transfer learning on the basis of the Mixture-of-Expert (MoE) architecture. However, the MoE-based method, which aims to project all information in the same feature space, cannot effectively deal with the complex relationships inherent among various scenarios and tasks, resulting in unsatisfactory performance. To tackle the problem, we propose a Hierarchical information extraction Network (HiNet) for multi-scenario and multi-task recommendation, which achieves hierarchical extraction based on coarse-to-fine knowledge transfer scheme. The multiple extraction layers of the hierarchical network enable the model to enhance the capability of transferring valuable information across scenarios while preserving specific features of scenarios and tasks. Furthermore, a novel scenario-aware attentive network module is proposed to model correlations between scenarios explicitly. Comprehensive experiments conducted on real-world industrial datasets from Meituan Meishi platform demonstrate that HiNet achieves a new state-of-the-art performance and significantly outperforms existing solutions. HiNet is currently fully deployed in two scenarios and has achieved 2.87% and 1.75% order quantity gain respectively.
Towards Generalized Open Information Extraction
Nov 29, 2022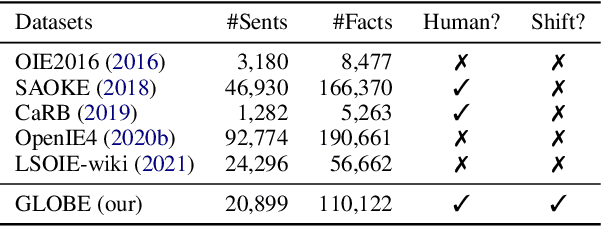
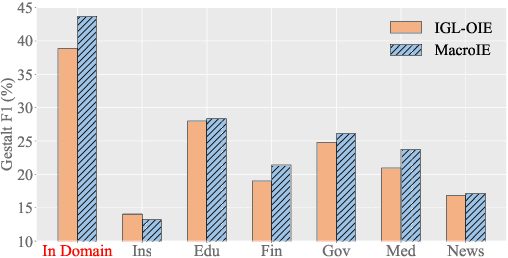
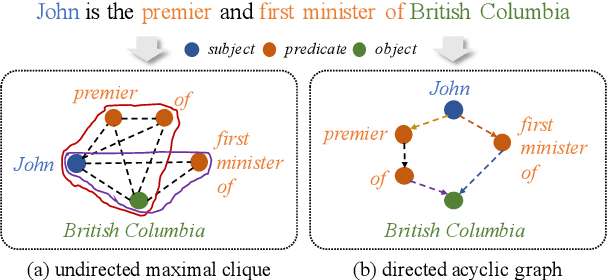
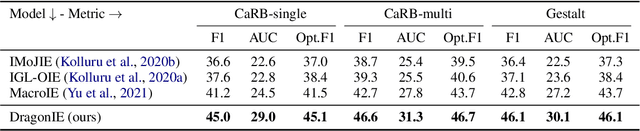
Open Information Extraction (OpenIE) facilitates the open-domain discovery of textual facts. However, the prevailing solutions evaluate OpenIE models on in-domain test sets aside from the training corpus, which certainly violates the initial task principle of domain-independence. In this paper, we propose to advance OpenIE towards a more realistic scenario: generalizing over unseen target domains with different data distributions from the source training domains, termed Generalized OpenIE. For this purpose, we first introduce GLOBE, a large-scale human-annotated multi-domain OpenIE benchmark, to examine the robustness of recent OpenIE models to domain shifts, and the relative performance degradation of up to 70% implies the challenges of generalized OpenIE. Then, we propose DragonIE, which explores a minimalist graph expression of textual fact: directed acyclic graph, to improve the OpenIE generalization. Extensive experiments demonstrate that DragonIE beats the previous methods in both in-domain and out-of-domain settings by as much as 6.0% in F1 score absolutely, but there is still ample room for improvement.
Dual-Stream Knowledge-Preserving Hashing for Unsupervised Video Retrieval
Oct 12, 2023Unsupervised video hashing usually optimizes binary codes by learning to reconstruct input videos. Such reconstruction constraint spends much effort on frame-level temporal context changes without focusing on video-level global semantics that are more useful for retrieval. Hence, we address this problem by decomposing video information into reconstruction-dependent and semantic-dependent information, which disentangles the semantic extraction from reconstruction constraint. Specifically, we first design a simple dual-stream structure, including a temporal layer and a hash layer. Then, with the help of semantic similarity knowledge obtained from self-supervision, the hash layer learns to capture information for semantic retrieval, while the temporal layer learns to capture the information for reconstruction. In this way, the model naturally preserves the disentangled semantics into binary codes. Validated by comprehensive experiments, our method consistently outperforms the state-of-the-arts on three video benchmarks.
Dual Defense: Adversarial, Traceable, and Invisible Robust Watermarking against Face Swapping
Oct 25, 2023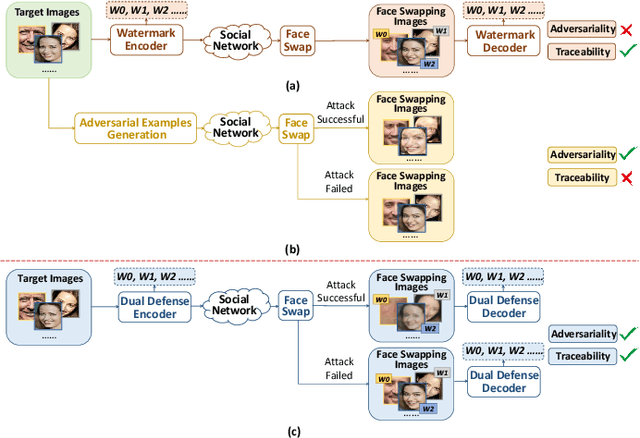
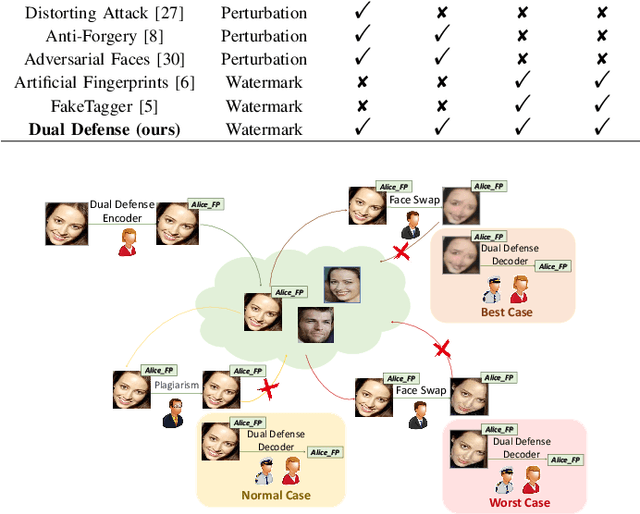
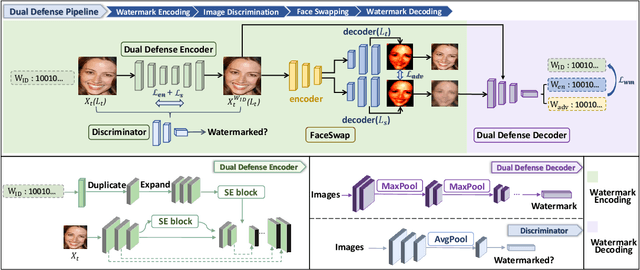
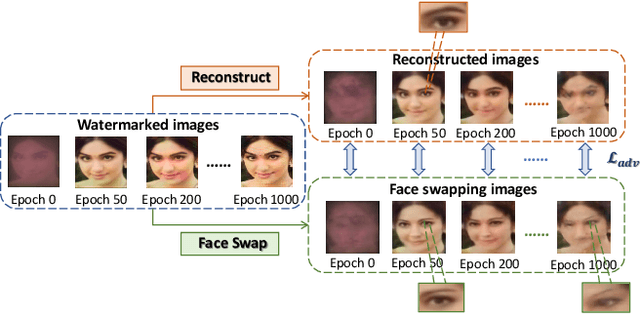
The malicious applications of deep forgery, represented by face swapping, have introduced security threats such as misinformation dissemination and identity fraud. While some research has proposed the use of robust watermarking methods to trace the copyright of facial images for post-event traceability, these methods cannot effectively prevent the generation of forgeries at the source and curb their dissemination. To address this problem, we propose a novel comprehensive active defense mechanism that combines traceability and adversariality, called Dual Defense. Dual Defense invisibly embeds a single robust watermark within the target face to actively respond to sudden cases of malicious face swapping. It disrupts the output of the face swapping model while maintaining the integrity of watermark information throughout the entire dissemination process. This allows for watermark extraction at any stage of image tracking for traceability. Specifically, we introduce a watermark embedding network based on original-domain feature impersonation attack. This network learns robust adversarial features of target facial images and embeds watermarks, seeking a well-balanced trade-off between watermark invisibility, adversariality, and traceability through perceptual adversarial encoding strategies. Extensive experiments demonstrate that Dual Defense achieves optimal overall defense success rates and exhibits promising universality in anti-face swapping tasks and dataset generalization ability. It maintains impressive adversariality and traceability in both original and robust settings, surpassing current forgery defense methods that possess only one of these capabilities, including CMUA-Watermark, Anti-Forgery, FakeTagger, or PGD methods.
SwG-former: Sliding-window Graph Convolutional Network Integrated with Conformer for Sound Event Localization and Detection
Oct 21, 2023Sound event localization and detection (SELD) is a joint task of sound event detection (SED) and direction of arrival (DoA) estimation. SED mainly relies on temporal dependencies to distinguish different sound classes, while DoA estimation depends on spatial correlations to estimate source directions. To jointly optimize two subtasks, the SELD system should extract spatial correlations and model temporal dependencies simultaneously. However, numerous models mainly extract spatial correlations and model temporal dependencies separately. In this paper, the interdependence of spatial-temporal information in audio signals is exploited for simultaneous extraction to enhance the model performance. In response, a novel graph representation leveraging graph convolutional network (GCN) in non-Euclidean space is developed to extract spatial-temporal information concurrently. A sliding-window graph (SwG) module is designed based on the graph representation. It exploits sliding-windows with different sizes to learn temporal context information and dynamically constructs graph vertices in the frequency-channel (F-C) domain to capture spatial correlations. Furthermore, as the cornerstone of message passing, a robust Conv2dAgg function is proposed and embedded into the SwG module to aggregate the features of neighbor vertices. To improve the performance of SELD in a natural spatial acoustic environment, a general and efficient SwG-former model is proposed by integrating the SwG module with the Conformer. It exhibits superior performance in comparison to recent advanced SELD models. To further validate the generality and efficiency of the SwG-former, it is seamlessly integrated into the event-independent network version 2 (EINV2) called SwG-EINV2. The SwG-EINV2 surpasses the state-of-the-art (SOTA) methods under the same acoustic environment.
Enhancing Argument Structure Extraction with Efficient Leverage of Contextual Information
Oct 08, 2023
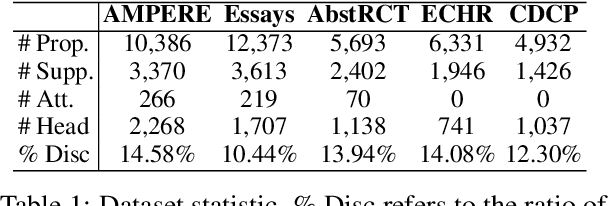
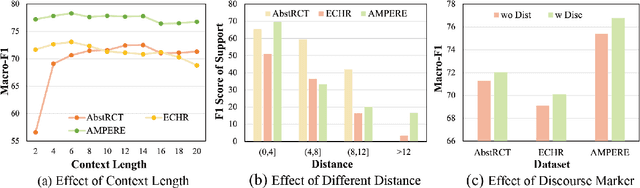
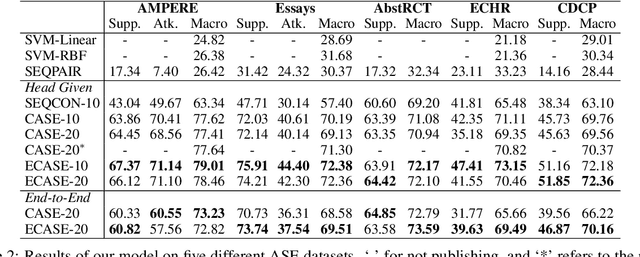
Argument structure extraction (ASE) aims to identify the discourse structure of arguments within documents. Previous research has demonstrated that contextual information is crucial for developing an effective ASE model. However, we observe that merely concatenating sentences in a contextual window does not fully utilize contextual information and can sometimes lead to excessive attention on less informative sentences. To tackle this challenge, we propose an Efficient Context-aware ASE model (ECASE) that fully exploits contextual information by enhancing modeling capacity and augmenting training data. Specifically, we introduce a sequence-attention module and distance-weighted similarity loss to aggregate contextual information and argumentative information. Additionally, we augment the training data by randomly masking discourse markers and sentences, which reduces the model's reliance on specific words or less informative sentences. Our experiments on five datasets from various domains demonstrate that our model achieves state-of-the-art performance. Furthermore, ablation studies confirm the effectiveness of each module in our model.
On Event Individuation for Document-Level Information Extraction
Dec 19, 2022
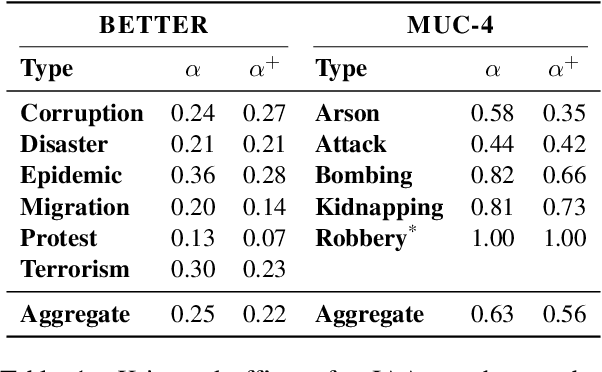

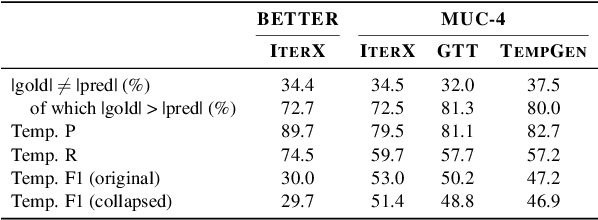
As information extraction (IE) systems have grown more capable at whole-document extraction, the classic task of \emph{template filling} has seen renewed interest as a benchmark for evaluating them. In this position paper, we call into question the suitability of template filling for this purpose. We argue that the task demands definitive answers to thorny questions of \emph{event individuation} -- the problem of distinguishing distinct events -- about which even human experts disagree. We show through annotation studies and error analysis that this raises concerns about the usefulness of template filling evaluation metrics, the quality of datasets for the task, and the ability of models to learn it. Finally, we consider possible solutions.
Product Attribute Value Extraction using Large Language Models
Oct 19, 2023E-commerce applications such as faceted product search or product comparison are based on structured product descriptions like attribute/value pairs. The vendors on e-commerce platforms do not provide structured product descriptions but describe offers using titles or descriptions. To process such offers, it is necessary to extract attribute/value pairs from textual product attributes. State-of-the-art attribute/value extraction techniques rely on pre-trained language models (PLMs), such as BERT. Two major drawbacks of these models for attribute/value extraction are that (i) the models require significant amounts of task-specific training data and (ii) the fine-tuned models face challenges in generalizing to attribute values not included in the training data. This paper explores the potential of large language models (LLMs) as a training data-efficient and robust alternative to PLM-based attribute/value extraction methods. We consider hosted LLMs, such as GPT-3.5 and GPT-4, as well as open-source LLMs based on Llama2. We evaluate the models in a zero-shot scenario and in a scenario where task-specific training data is available. In the zero-shot scenario, we compare various prompt designs for representing information about the target attributes of the extraction. In the scenario with training data, we investigate (i) the provision of example attribute values, (ii) the selection of in-context demonstrations, and (iii) the fine-tuning of GPT-3.5. Our experiments show that GPT-4 achieves an average F1-score of 85% on the two evaluation datasets while the best PLM-based techniques perform on average 5% worse using the same amount of training data. GPT-4 achieves a 10% higher F1-score than the best open-source LLM. The fine-tuned GPT-3.5 model reaches a similar performance as GPT-4 while being significantly more cost-efficient.
Be Careful What You Smooth For: Label Smoothing Can Be a Privacy Shield but Also a Catalyst for Model Inversion Attacks
Oct 10, 2023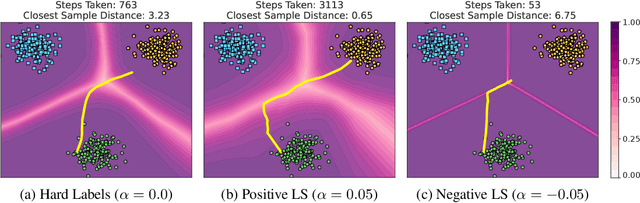
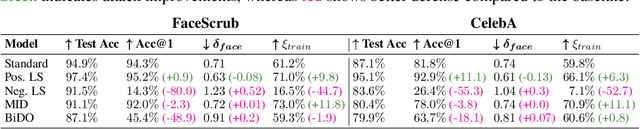
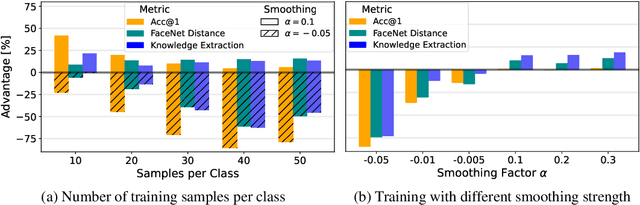
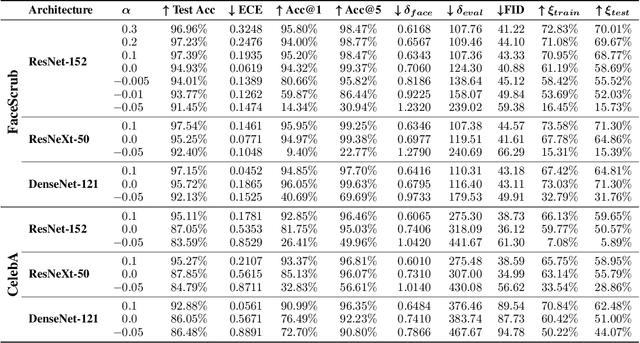
Label smoothing -- using softened labels instead of hard ones -- is a widely adopted regularization method for deep learning, showing diverse benefits such as enhanced generalization and calibration. Its implications for preserving model privacy, however, have remained unexplored. To fill this gap, we investigate the impact of label smoothing on model inversion attacks (MIAs), which aim to generate class-representative samples by exploiting the knowledge encoded in a classifier, thereby inferring sensitive information about its training data. Through extensive analyses, we uncover that traditional label smoothing fosters MIAs, thereby increasing a model's privacy leakage. Even more, we reveal that smoothing with negative factors counters this trend, impeding the extraction of class-related information and leading to privacy preservation, beating state-of-the-art defenses. This establishes a practical and powerful novel way for enhancing model resilience against MIAs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge