"Information Extraction": models, code, and papers
Rethinking E-Commerce Search
Dec 06, 2023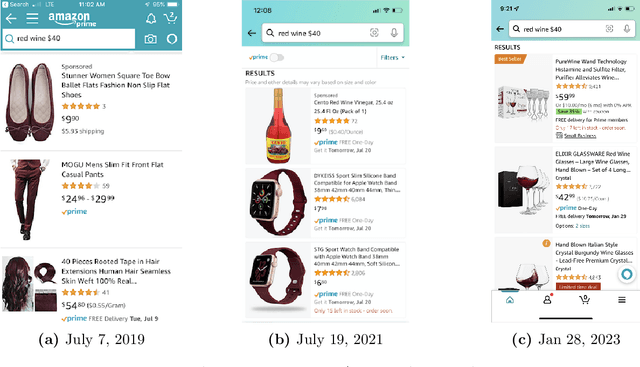
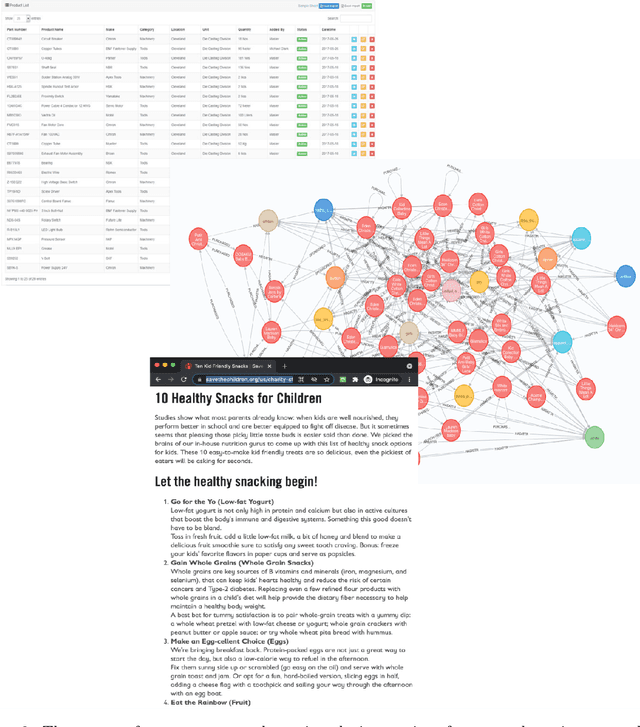
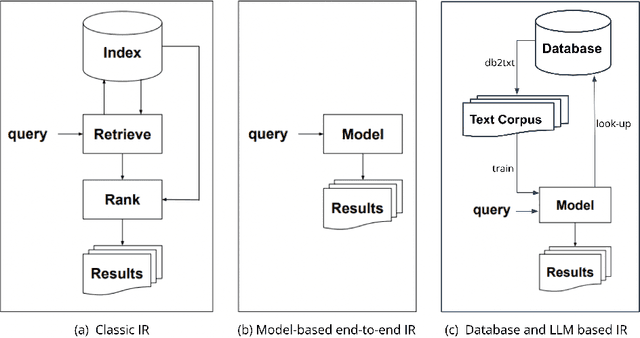

E-commerce search and recommendation usually operate on structured data such as product catalogs and taxonomies. However, creating better search and recommendation systems often requires a large variety of unstructured data including customer reviews and articles on the web. Traditionally, the solution has always been converting unstructured data into structured data through information extraction, and conducting search over the structured data. However, this is a costly approach that often has low quality. In this paper, we envision a solution that does entirely the opposite. Instead of converting unstructured data (web pages, customer reviews, etc) to structured data, we instead convert structured data (product inventory, catalogs, taxonomies, etc) into textual data, which can be easily integrated into the text corpus that trains LLMs. Then, search and recommendation can be performed through a Q/A mechanism through an LLM instead of using traditional information retrieval methods over structured data.
MT4CrossOIE: Multi-stage Tuning for Cross-lingual Open Information Extraction
Aug 12, 2023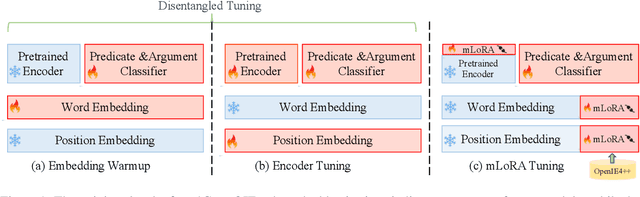

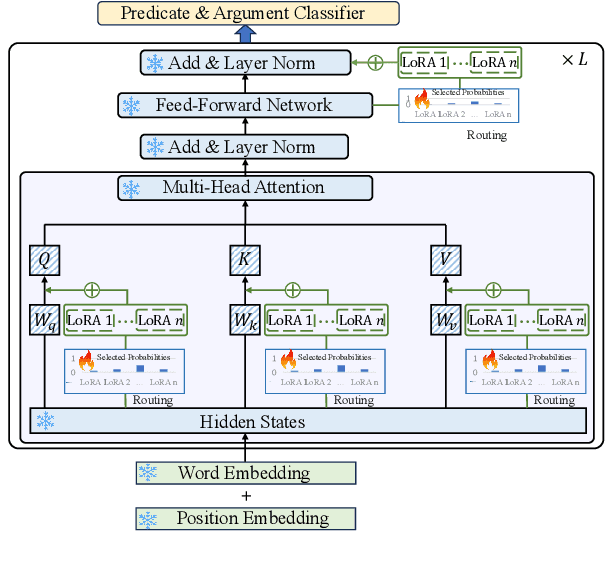
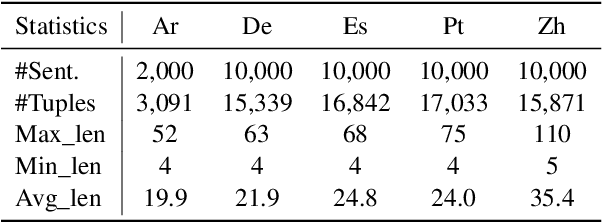
Cross-lingual open information extraction aims to extract structured information from raw text across multiple languages. Previous work uses a shared cross-lingual pre-trained model to handle the different languages but underuses the potential of the language-specific representation. In this paper, we propose an effective multi-stage tuning framework called MT4CrossIE, designed for enhancing cross-lingual open information extraction by injecting language-specific knowledge into the shared model. Specifically, the cross-lingual pre-trained model is first tuned in a shared semantic space (e.g., embedding matrix) in the fixed encoder and then other components are optimized in the second stage. After enough training, we freeze the pre-trained model and tune the multiple extra low-rank language-specific modules using mixture-of-LoRAs for model-based cross-lingual transfer. In addition, we leverage two-stage prompting to encourage the large language model (LLM) to annotate the multi-lingual raw data for data-based cross-lingual transfer. The model is trained with multi-lingual objectives on our proposed dataset OpenIE4++ by combing the model-based and data-based transfer techniques. Experimental results on various benchmarks emphasize the importance of aggregating multiple plug-in-and-play language-specific modules and demonstrate the effectiveness of MT4CrossIE in cross-lingual OIE\footnote{\url{https://github.com/CSJianYang/Multilingual-Multimodal-NLP}}.
Enhanced Information Extraction from Cylindrical Visual-Tactile Sensors via Image Fusion
Nov 07, 2023Vision-based tactile sensors equipped with planar contact structures acquire the shape, force, and motion states of objects in contact. The limited planar contact area presents a challenge in acquiring information about larger target objects. In contrast, vision-based tactile sensors with cylindrical contact structures could extend the contact area by rolling, which can acquire much tactile information that exceeds the sensing projection area in a single contact. However, the tactile data acquired by cylindrical structures does not consistently correspond to the same depth level. Therefore, stitching and analyzing the data in an extended contact area is a challenging problem. In this work, we propose an image fusion method based on cylindrical vision-based tactile sensors. The method takes advantage of the changing characteristics of the contact depth of cylindrical structures, extracts the effective information of different contact depths in the frequency domain, and performs differential fusion for the information characteristics. The results show that in object contact confronting an area larger than single sensing, the images fused with our proposed method have higher information and structural similarity compared with the method of stitching based on motion distance sampling. Meanwhile, it is robust to sampling time. We complement this method with a deep neural network to illustrate its potential for fusing and recognizing object contact information using cylindrical vision-based tactile sensors.
Distribution-aware Interactive Attention Network and Large-scale Cloud Recognition Benchmark on FY-4A Satellite Image
Jan 06, 2024Accurate cloud recognition and warning are crucial for various applications, including in-flight support, weather forecasting, and climate research. However, recent deep learning algorithms have predominantly focused on detecting cloud regions in satellite imagery, with insufficient attention to the specificity required for accurate cloud recognition. This limitation inspired us to develop the novel FY-4A-Himawari-8 (FYH) dataset, which includes nine distinct cloud categories and uses precise domain adaptation methods to align 70,419 image-label pairs in terms of projection, temporal resolution, and spatial resolution, thereby facilitating the training of supervised deep learning networks. Given the complexity and diversity of cloud formations, we have thoroughly analyzed the challenges inherent to cloud recognition tasks, examining the intricate characteristics and distribution of the data. To effectively address these challenges, we designed a Distribution-aware Interactive-Attention Network (DIAnet), which preserves pixel-level details through a high-resolution branch and a parallel multi-resolution cross-branch. We also integrated a distribution-aware loss (DAL) to mitigate the imbalance across cloud categories. An Interactive Attention Module (IAM) further enhances the robustness of feature extraction combined with spatial and channel information. Empirical evaluations on the FYH dataset demonstrate that our method outperforms other cloud recognition networks, achieving superior performance in terms of mean Intersection over Union (mIoU). The code for implementing DIAnet is available at https://github.com/icey-zhang/DIAnet.
AI-driven Structure Detection and Information Extraction from Historical Cadastral Maps (Early 19th Century Franciscean Cadastre in the Province of Styria) and Current High-resolution Satellite and Aerial Imagery for Remote Sensing
Dec 08, 2023Cadastres from the 19th century are a complex as well as rich source for historians and archaeologists, whose use presents them with great challenges. For archaeological and historical remote sensing, we have trained several Deep Learning models, CNNs as well as Vision Transformers, to extract large-scale data from this knowledge representation. We present the principle results of our work here and we present a the demonstrator of our browser-based tool that allows researchers and public stakeholders to quickly identify spots that featured buildings in the 19th century Franciscean Cadastre. The tool not only supports scholars and fellow researchers in building a better understanding of the settlement history of the region of Styria, it also helps public administration and fellow citizens to swiftly identify areas of heightened sensibility with regard to the cultural heritage of the region.
MobileUtr: Revisiting the relationship between light-weight CNN and Transformer for efficient medical image segmentation
Dec 04, 2023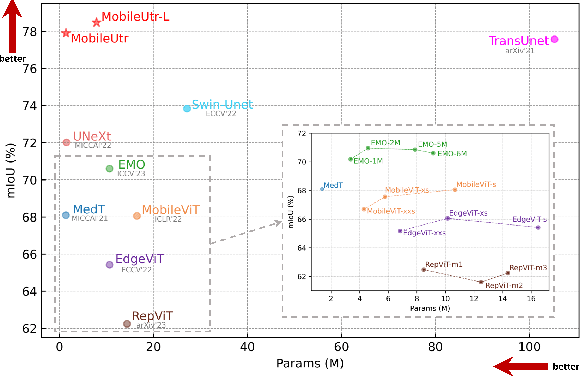
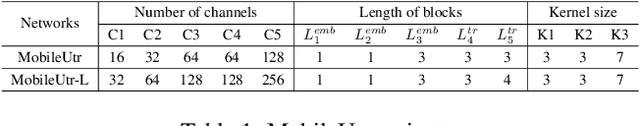
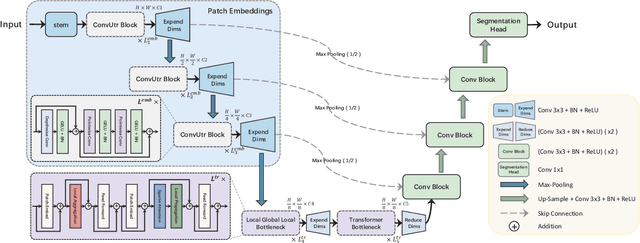
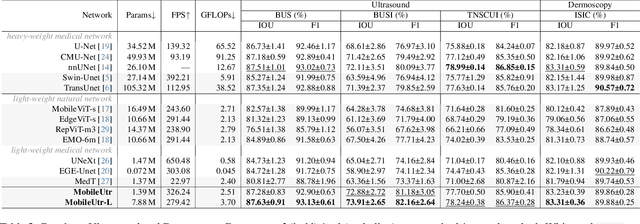
Due to the scarcity and specific imaging characteristics in medical images, light-weighting Vision Transformers (ViTs) for efficient medical image segmentation is a significant challenge, and current studies have not yet paid attention to this issue. This work revisits the relationship between CNNs and Transformers in lightweight universal networks for medical image segmentation, aiming to integrate the advantages of both worlds at the infrastructure design level. In order to leverage the inductive bias inherent in CNNs, we abstract a Transformer-like lightweight CNNs block (ConvUtr) as the patch embeddings of ViTs, feeding Transformer with denoised, non-redundant and highly condensed semantic information. Moreover, an adaptive Local-Global-Local (LGL) block is introduced to facilitate efficient local-to-global information flow exchange, maximizing Transformer's global context information extraction capabilities. Finally, we build an efficient medical image segmentation model (MobileUtr) based on CNN and Transformer. Extensive experiments on five public medical image datasets with three different modalities demonstrate the superiority of MobileUtr over the state-of-the-art methods, while boasting lighter weights and lower computational cost. Code is available at https://github.com/FengheTan9/MobileUtr.
Improving Open Information Extraction with Large Language Models: A Study on Demonstration Uncertainty
Sep 07, 2023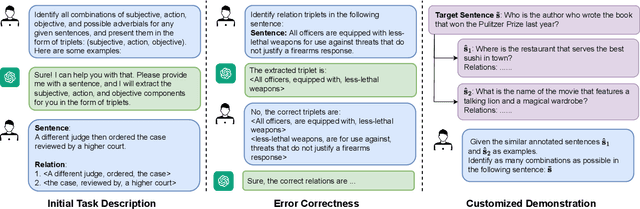
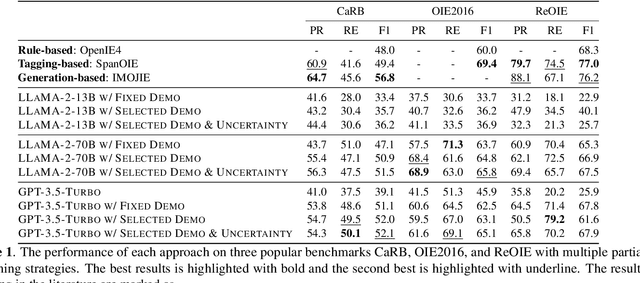

Open Information Extraction (OIE) task aims at extracting structured facts from unstructured text, typically in the form of (subject, relation, object) triples. Despite the potential of large language models (LLMs) like ChatGPT as a general task solver, they lag behind state-of-the-art (supervised) methods in OIE tasks due to two key issues. First, LLMs struggle to distinguish irrelevant context from relevant relations and generate structured output due to the restrictions on fine-tuning the model. Second, LLMs generates responses autoregressively based on probability, which makes the predicted relations lack confidence. In this paper, we assess the capabilities of LLMs in improving the OIE task. Particularly, we propose various in-context learning strategies to enhance LLM's instruction-following ability and a demonstration uncertainty quantification module to enhance the confidence of the generated relations. Our experiments on three OIE benchmark datasets show that our approach holds its own against established supervised methods, both quantitatively and qualitatively.
Self-Supervised Disentangled Representation Learning for Robust Target Speech Extraction
Dec 16, 2023Speech signals are inherently complex as they encompass both global acoustic characteristics and local semantic information. However, in the task of target speech extraction, certain elements of global and local semantic information in the reference speech, which are irrelevant to speaker identity, can lead to speaker confusion within the speech extraction network. To overcome this challenge, we propose a self-supervised disentangled representation learning method. Our approach tackles this issue through a two-phase process, utilizing a reference speech encoding network and a global information disentanglement network to gradually disentangle the speaker identity information from other irrelevant factors. We exclusively employ the disentangled speaker identity information to guide the speech extraction network. Moreover, we introduce the adaptive modulation Transformer to ensure that the acoustic representation of the mixed signal remains undisturbed by the speaker embeddings. This component incorporates speaker embeddings as conditional information, facilitating natural and efficient guidance for the speech extraction network. Experimental results substantiate the effectiveness of our meticulously crafted approach, showcasing a substantial reduction in the likelihood of speaker confusion.
Wasserstein Nonnegative Tensor Factorization with Manifold Regularization
Jan 03, 2024Nonnegative tensor factorization (NTF) has become an important tool for feature extraction and part-based representation with preserved intrinsic structure information from nonnegative high-order data. However, the original NTF methods utilize Euclidean or Kullback-Leibler divergence as the loss function which treats each feature equally leading to the neglect of the side-information of features. To utilize correlation information of features and manifold information of samples, we introduce Wasserstein manifold nonnegative tensor factorization (WMNTF), which minimizes the Wasserstein distance between the distribution of input tensorial data and the distribution of reconstruction. Although some researches about Wasserstein distance have been proposed in nonnegative matrix factorization (NMF), they ignore the spatial structure information of higher-order data. We use Wasserstein distance (a.k.a Earth Mover's distance or Optimal Transport distance) as a metric and add a graph regularizer to a latent factor. Experimental results demonstrate the effectiveness of the proposed method compared with other NMF and NTF methods.
A Temporal Filter to Extract Doped Conducting Polymer Information Features from an Electronic Nose
Jan 01, 2024Identifying relevant machine-learning features for multi-sensing platforms is both an applicative limitation to recognize environments and a necessity to interpret the physical relevance of transducers' complementarity in their information processing. Particularly for long acquisitions, feature extraction must be fully automatized without human intervention and resilient to perturbations without increasing significantly the computational cost of a classifier. In this study, we investigate on the relative resistance and current modulation of a 24-dimensional conductimetric electronic nose, which uses the exponential moving average as a floating reference in a low-cost information descriptor for environment recognition. In particular, we identified that depending on the structure of a linear classifier, the 'modema' descriptor is optimized for different material sensing elements' contributions to classify information patterns. The low-pass filtering optimization leads to opposite behaviors between unsupervised and supervised learning: the latter one favors longer integration of the reference, allowing to recognize five different classes over 90%, while the first one prefers using the latest events as its reference to clusterize patterns by environment nature. Its electronic implementation shall greatly diminish the computational requirements of conductimetric electronic noses for on-board environment recognition without human supervision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge