Isaiah Onando Mulang'
Investigating Expressiveness of Transformer in Spectral Domain for Graphs
Jan 27, 2022
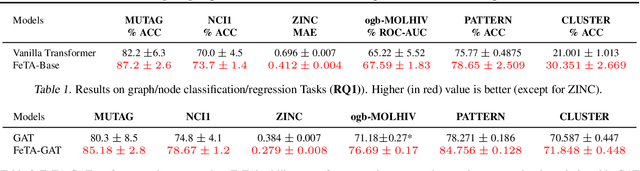
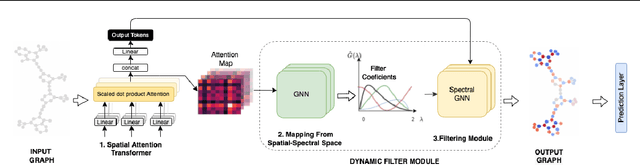
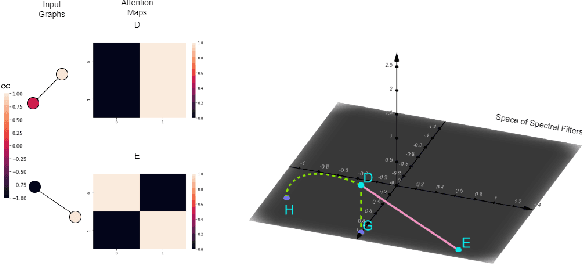
Abstract:Transformers have been proven to be inadequate for graph representation learning. To understand this inadequacy, there is need to investigate if spectral analysis of transformer will reveal insights on its expressive power. Similar studies already established that spectral analysis of Graph neural networks (GNNs) provides extra perspectives on their expressiveness. In this work, we systematically study and prove the link between the spatial and spectral domain in the realm of the transformer. We further provide a theoretical analysis that the spatial attention mechanism in the transformer cannot effectively capture the desired frequency response, thus, inherently limiting its expressiveness in spectral space. Therefore, we propose FeTA, a framework that aims to perform attention over the entire graph spectrum analogous to the attention in spatial space. Empirical results suggest that FeTA provides homogeneous performance gain against vanilla transformer across all tasks on standard benchmarks and can easily be extended to GNN based models with low-pass characteristics (e.g., GAT). Furthermore, replacing the vanilla transformer model with FeTA in recently proposed position encoding schemes has resulted in comparable or better performance than transformer and GNN baselines.
Sparsity-based Feature Selection for Anomalous Subgroup Discovery
Jan 06, 2022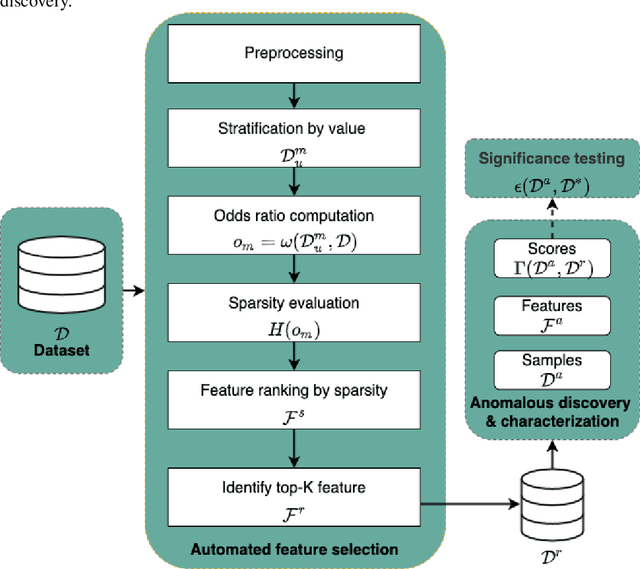
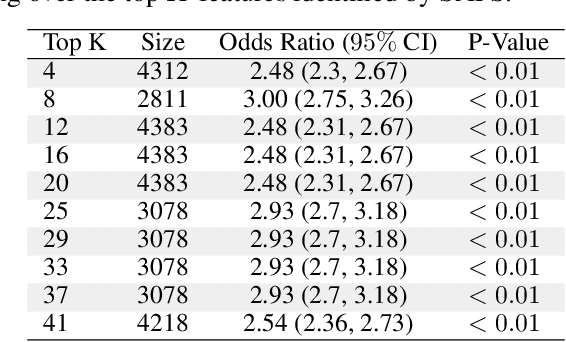
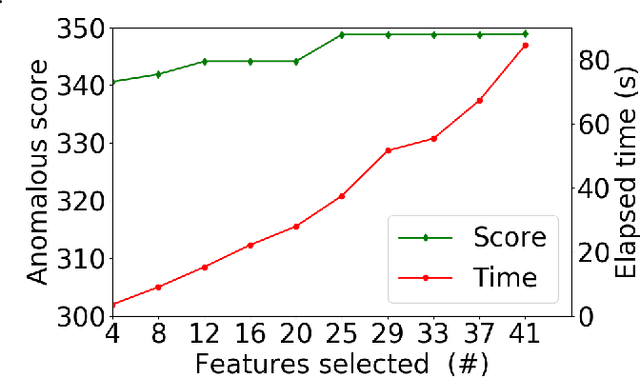
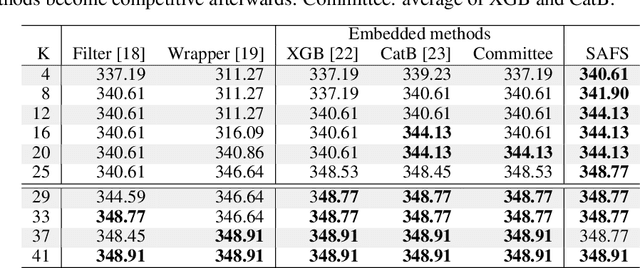
Abstract:Anomalous pattern detection aims to identify instances where deviation from normalcy is evident, and is widely applicable across domains. Multiple anomalous detection techniques have been proposed in the state of the art. However, there is a common lack of a principled and scalable feature selection method for efficient discovery. Existing feature selection techniques are often conducted by optimizing the performance of prediction outcomes rather than its systemic deviations from the expected. In this paper, we proposed a sparsity-based automated feature selection (SAFS) framework, which encodes systemic outcome deviations via the sparsity of feature-driven odds ratios. SAFS is a model-agnostic approach with usability across different discovery techniques. SAFS achieves more than $3\times$ reduction in computation time while maintaining detection performance when validated on publicly available critical care dataset. SAFS also results in a superior performance when compared against multiple baselines for feature selection.
Post-discovery Analysis of Anomalous Subsets
Nov 23, 2021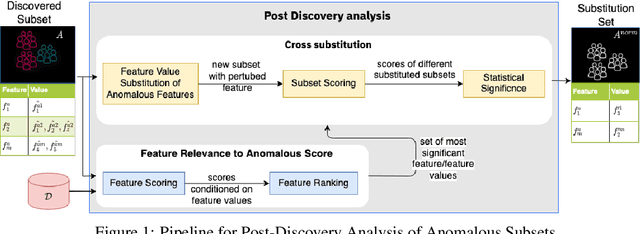
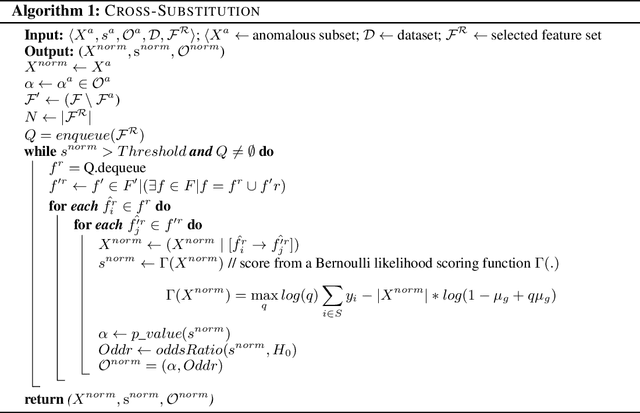
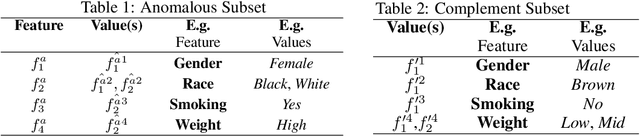
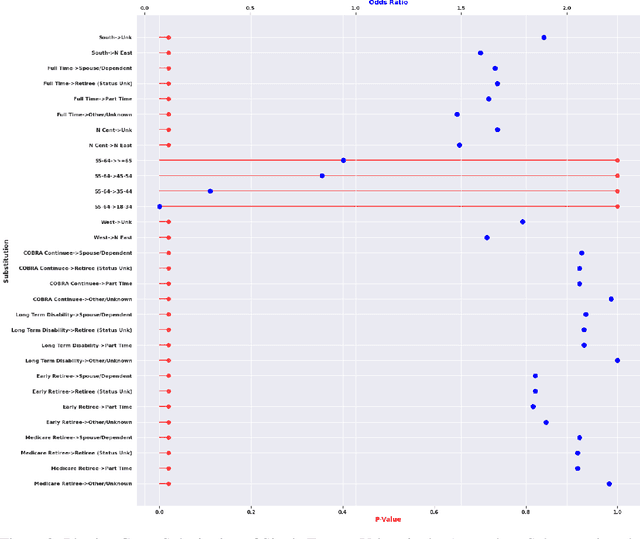
Abstract:Analyzing the behaviour of a population in response to disease and interventions is critical to unearth variability in healthcare as well as understand sub-populations that require specialized attention, but also to assist in designing future interventions. Two aspects become very essential in such analysis namely: i) Discovery of differentiating patterns exhibited by sub-populations, and ii) Characterization of the identified subpopulations. For the discovery phase, an array of approaches in the anomalous pattern detection literature have been employed to reveal differentiating patterns, especially to identify anomalous subgroups. However, these techniques are limited to describing the anomalous subgroups and offer little in form of insightful characterization, thereby limiting interpretability and understanding of these data-driven techniques in clinical practices. In this work, we propose an analysis of differentiated output (rather than discovery) and quantify anomalousness similarly to the counter-factual setting. To this end we design an approach to perform post-discovery analysis of anomalous subsets, in which we initially identify the most important features on the anomalousness of the subsets, then by perturbation, the approach seeks to identify the least number of changes necessary to lose anomalousness. Our approach is presented and the evaluation results on the 2019 MarketScan Commercial Claims and Medicare data, show that extra insights can be obtained by extrapolated examination of the identified subgroups.
Automated Supervised Feature Selection for Differentiated Patterns of Care
Nov 05, 2021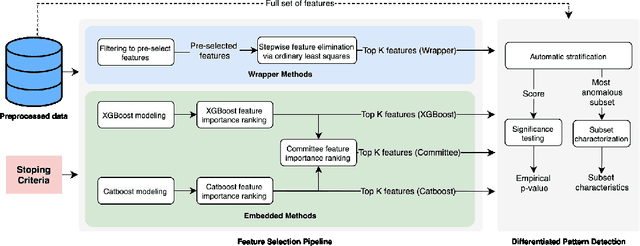
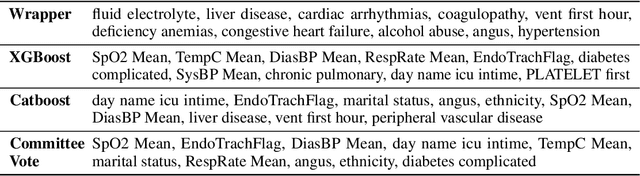
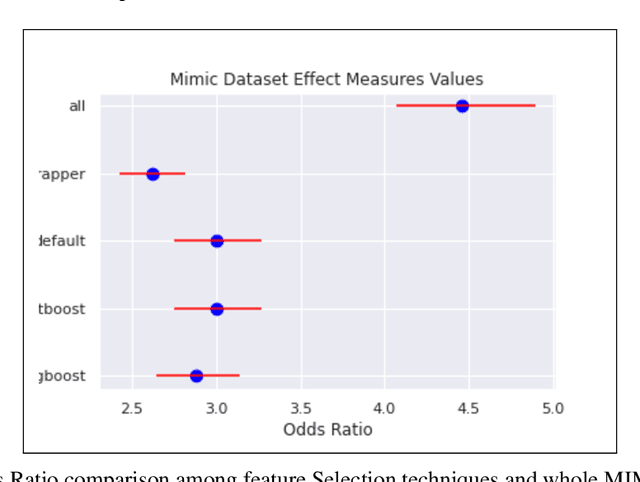
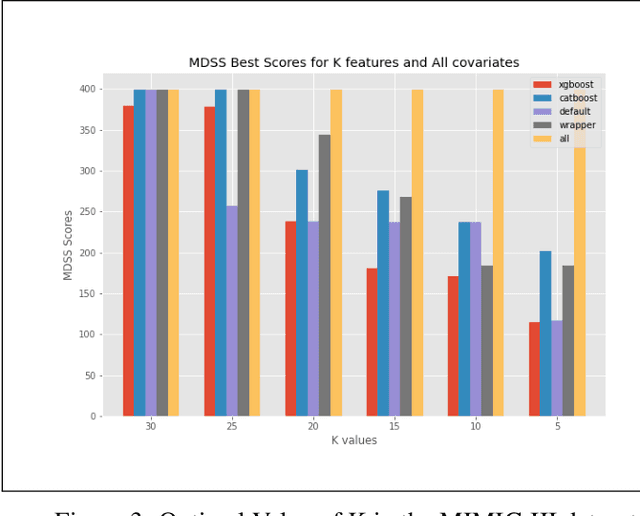
Abstract:An automated feature selection pipeline was developed using several state-of-the-art feature selection techniques to select optimal features for Differentiating Patterns of Care (DPOC). The pipeline included three types of feature selection techniques; Filters, Wrappers and Embedded methods to select the top K features. Five different datasets with binary dependent variables were used and their different top K optimal features selected. The selected features were tested in the existing multi-dimensional subset scanning (MDSS) where the most anomalous subpopulations, most anomalous subsets, propensity scores, and effect of measures were recorded to test their performance. This performance was compared with four similar metrics gained after using all covariates in the dataset in the MDSS pipeline. We found out that despite the different feature selection techniques used, the data distribution is key to note when determining the technique to use.
Ranking Facts for Explaining Answers to Elementary Science Questions
Oct 18, 2021
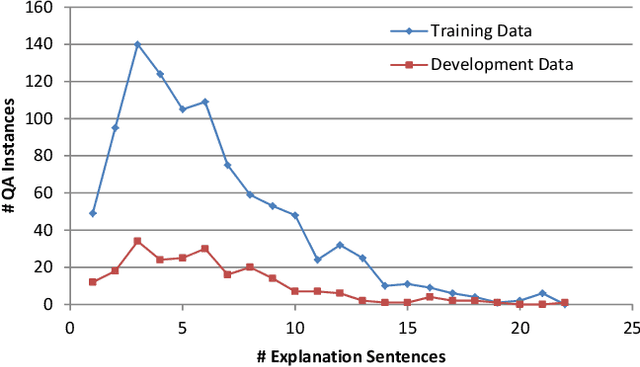

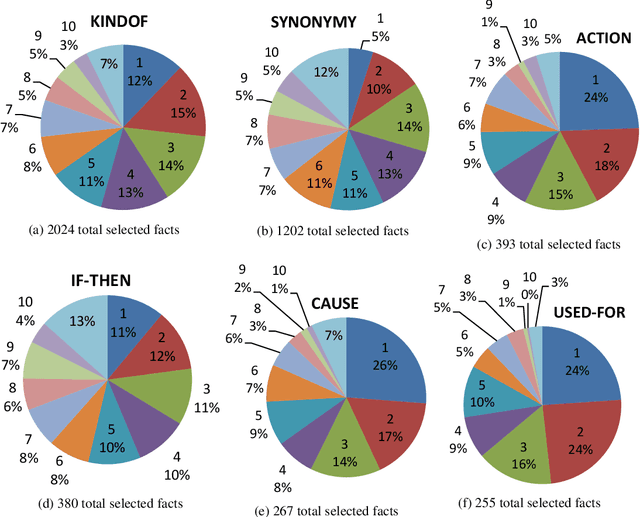
Abstract:In multiple-choice exams, students select one answer from among typically four choices and can explain why they made that particular choice. Students are good at understanding natural language questions and based on their domain knowledge can easily infer the question's answer by 'connecting the dots' across various pertinent facts. Considering automated reasoning for elementary science question answering, we address the novel task of generating explanations for answers from human-authored facts. For this, we examine the practically scalable framework of feature-rich support vector machines leveraging domain-targeted, hand-crafted features. Explanations are created from a human-annotated set of nearly 5,000 candidate facts in the WorldTree corpus. Our aim is to obtain better matches for valid facts of an explanation for the correct answer of a question over the available fact candidates. To this end, our features offer a comprehensive linguistic and semantic unification paradigm. The machine learning problem is the preference ordering of facts, for which we test pointwise regression versus pairwise learning-to-rank. Our contributions are: (1) a case study in which two preference ordering approaches are systematically compared; (2) it is a practically competent approach that can outperform some variants of BERT-based reranking models; and (3) the human-engineered features make it an interpretable machine learning model for the task.
KGPool: Dynamic Knowledge Graph Context Selection for Relation Extraction
Jun 06, 2021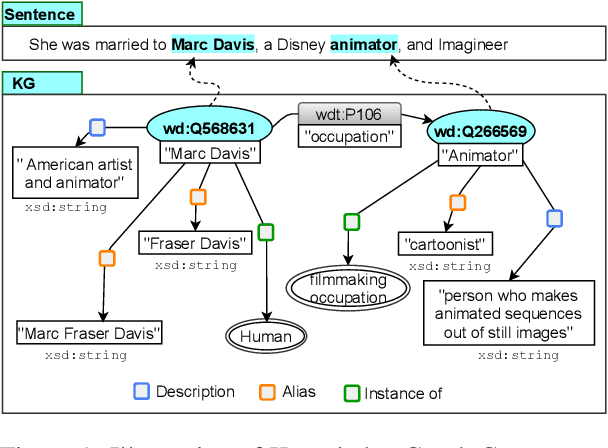
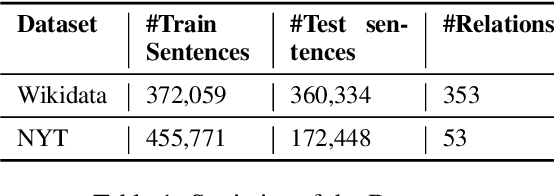
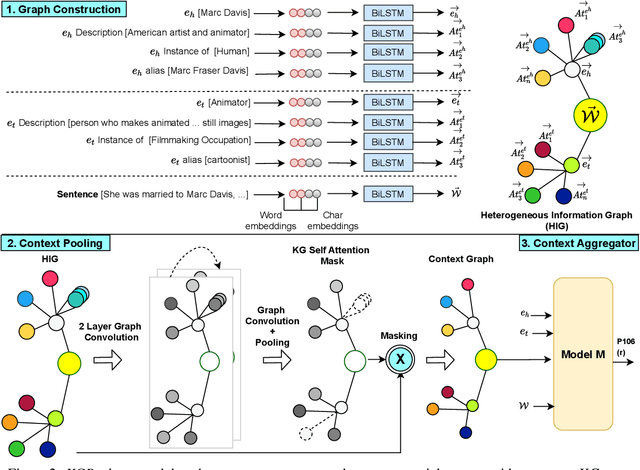
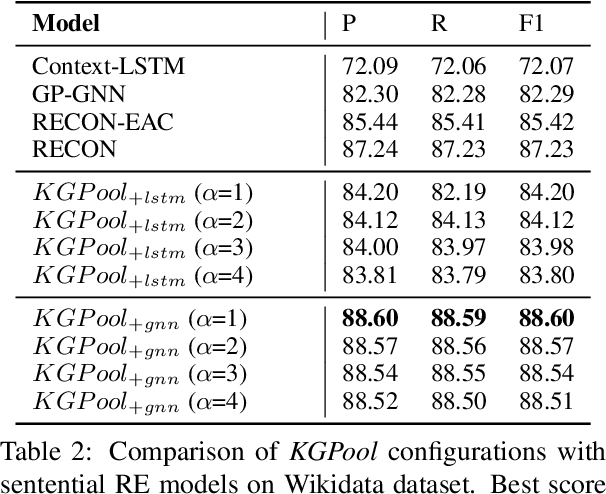
Abstract:We present a novel method for relation extraction (RE) from a single sentence, mapping the sentence and two given entities to a canonical fact in a knowledge graph (KG). Especially in this presumed sentential RE setting, the context of a single sentence is often sparse. This paper introduces the KGPool method to address this sparsity, dynamically expanding the context with additional facts from the KG. It learns the representation of these facts (entity alias, entity descriptions, etc.) using neural methods, supplementing the sentential context. Unlike existing methods that statically use all expanded facts, KGPool conditions this expansion on the sentence. We study the efficacy of KGPool by evaluating it with different neural models and KGs (Wikidata and NYT Freebase). Our experimental evaluation on standard datasets shows that by feeding the KGPool representation into a Graph Neural Network, the overall method is significantly more accurate than state-of-the-art methods.
CHOLAN: A Modular Approach for Neural Entity Linking on Wikipedia and Wikidata
Feb 08, 2021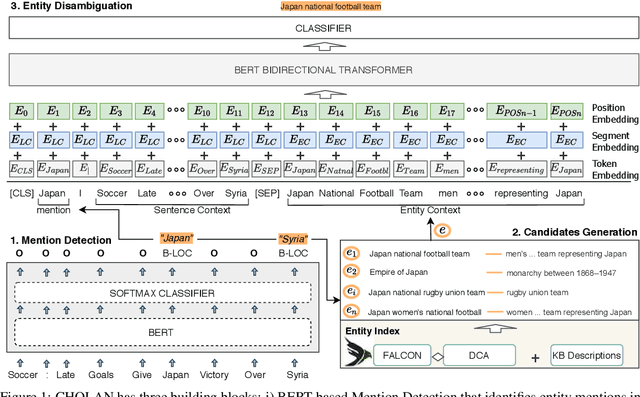
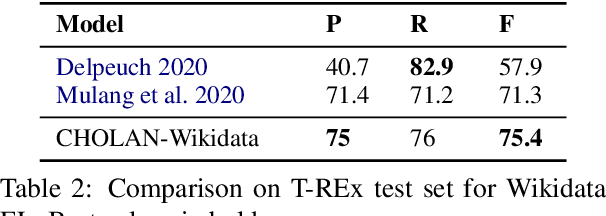
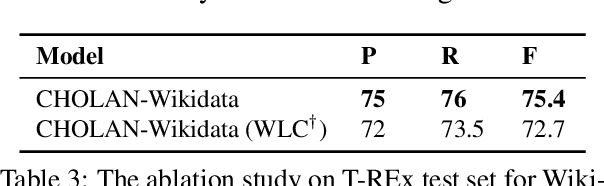
Abstract:In this paper, we propose CHOLAN, a modular approach to target end-to-end entity linking (EL) over knowledge bases. CHOLAN consists of a pipeline of two transformer-based models integrated sequentially to accomplish the EL task. The first transformer model identifies surface forms (entity mentions) in a given text. For each mention, a second transformer model is employed to classify the target entity among a predefined candidates list. The latter transformer is fed by an enriched context captured from the sentence (i.e. local context), and entity description gained from Wikipedia. Such external contexts have not been used in the state of the art EL approaches. Our empirical study was conducted on two well-known knowledge bases (i.e., Wikidata and Wikipedia). The empirical results suggest that CHOLAN outperforms state-of-the-art approaches on standard datasets such as CoNLL-AIDA, MSNBC, AQUAINT, ACE2004, and T-REx.
RECON: Relation Extraction using Knowledge Graph Context in a Graph Neural Network
Sep 18, 2020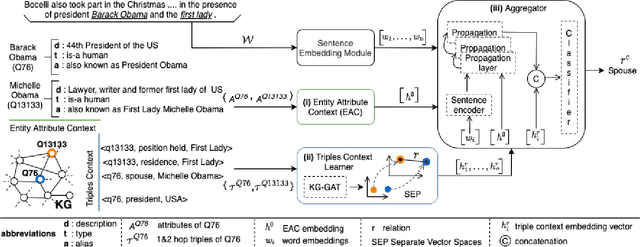
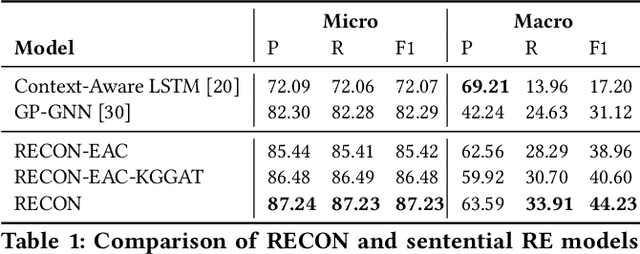
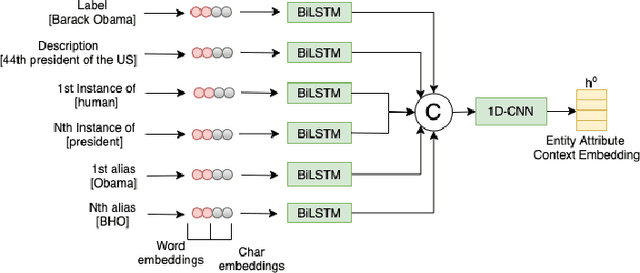
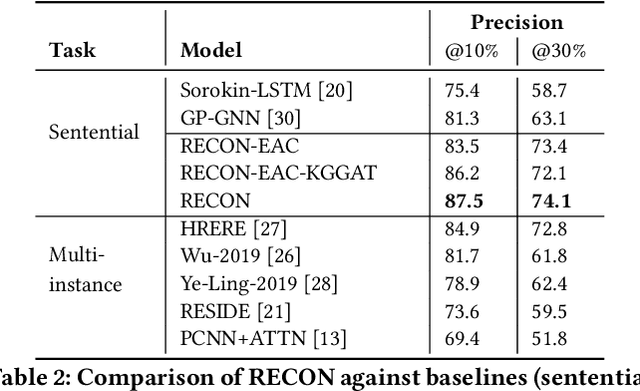
Abstract:In this paper, we present a novel method named RECON, that automatically identifies relations in a sentence (sentential relation extraction) and aligns to a knowledge graph (KG). RECON uses a graph neural network to learn representations of both the sentence as well as facts stored in a KG, improving the overall extraction quality. These facts, including entity attributes (label, alias, description, instance-of) and factual triples, have not been collectively used in the state of the art methods. We evaluate the effect of various forms of representing the KG context on the performance of RECON. The empirical evaluation on two standard relation extraction datasets shows that RECON significantly outperforms all state of the art methods on NYT Freebase and Wikidata datasets. RECON reports 87.23 F1 score (Vs 82.29 baseline) on Wikidata dataset whereas on NYT Freebase, reported values are 87.5(P@10) and 74.1(P@30) compared to the previous baseline scores of 81.3(P@10) and 63.1(P@30).
Evaluating the Impact of Knowledge Graph Context on Entity Disambiguation Models
Aug 30, 2020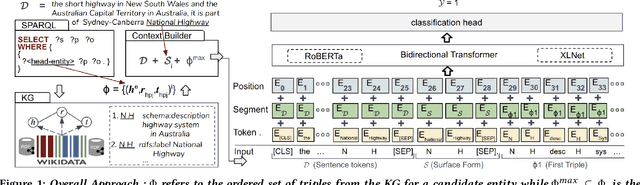
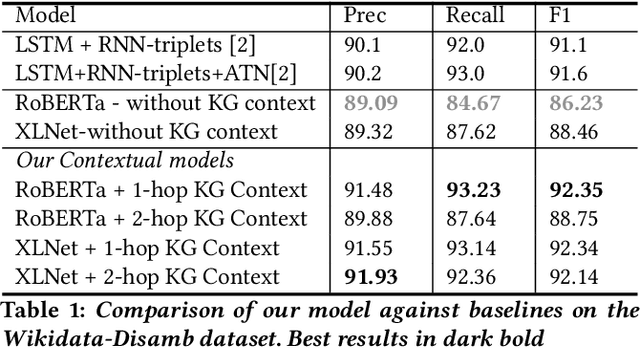

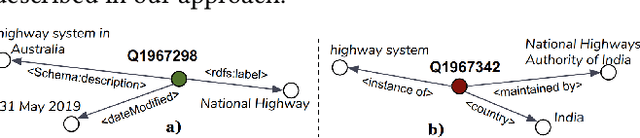
Abstract:Pretrained Transformer models have emerged as state-of-the-art approaches that learn contextual information from text to improve the performance of several NLP tasks. These models, albeit powerful, still require specialized knowledge in specific scenarios. In this paper, we argue that context derived from a knowledge graph (in our case: Wikidata) provides enough signals to inform pretrained transformer models and improve their performance for named entity disambiguation (NED) on Wikidata KG. We further hypothesize that our proposed KG context can be standardized for Wikipedia, and we evaluate the impact of KG context on state-of-the-art NED model for the Wikipedia knowledge base. Our empirical results validate that the proposed KG context can be generalized (for Wikipedia), and providing KG context in transformer architectures considerably outperforms the existing baselines, including the vanilla transformer models.
* to appear in proceedings of CIKM 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge