Gopinath Chennupati
On Mixup Training: Improved Calibration and Predictive Uncertainty for Deep Neural Networks
May 27, 2019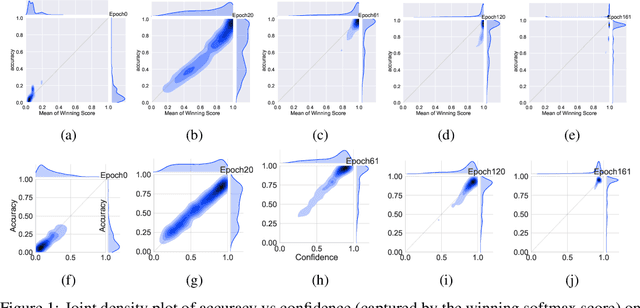
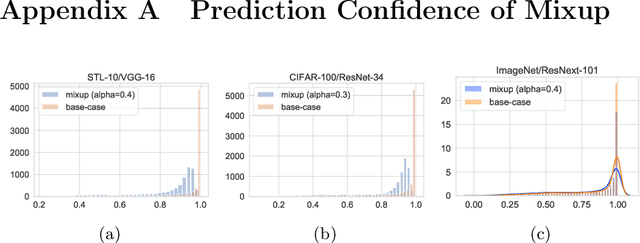
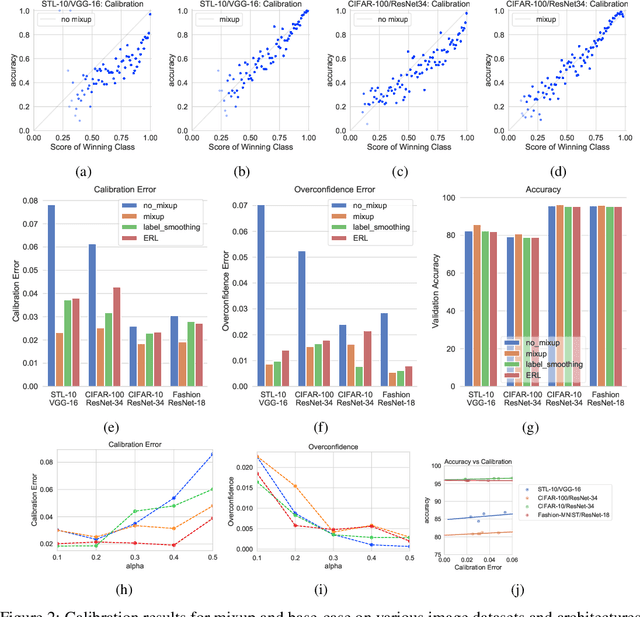
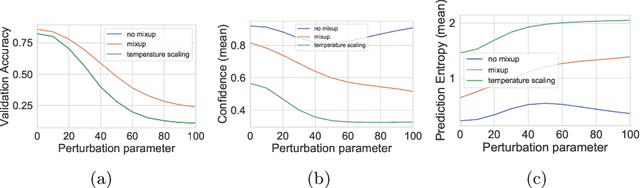
Abstract:Mixup~\cite{zhang2017mixup} is a recently proposed method for training deep neural networks where additional samples are generated during training by convexly combining random pairs of images and their associated labels. While simple to implement, it has shown to be a surprisingly effective method of data augmentation for image classification; DNNs trained with mixup show noticeable gains in classification performance on a number of image classification benchmarks. In this work, we discuss a hitherto untouched aspect of mixup training -- the calibration and predictive uncertainty of models trained with mixup. We find that DNNs trained with mixup are significantly better calibrated -- i.e., the predicted softmax scores are much better indicators of the actual likelihood of a correct prediction -- than DNNs trained in the regular fashion. We conduct experiments on a number of image classification architectures and datasets -- including large-scale datasets like ImageNet -- and find this to be the case. Additionally, we find that merely mixing features does not result in the same calibration benefit and that the label smoothing in mixup training plays a significant role in improving calibration. Finally, we also observe that mixup-trained DNNs are less prone to over-confident predictions on out-of-distribution and random-noise data. We conclude that the typical overconfidence seen in neural networks, even on in-distribution data is likely a consequence of training with hard labels, suggesting that mixup training be employed for classification tasks where predictive uncertainty is a significant concern.
Combating Label Noise in Deep Learning Using Abstention
May 27, 2019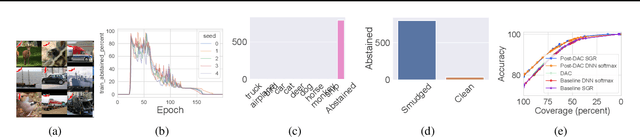
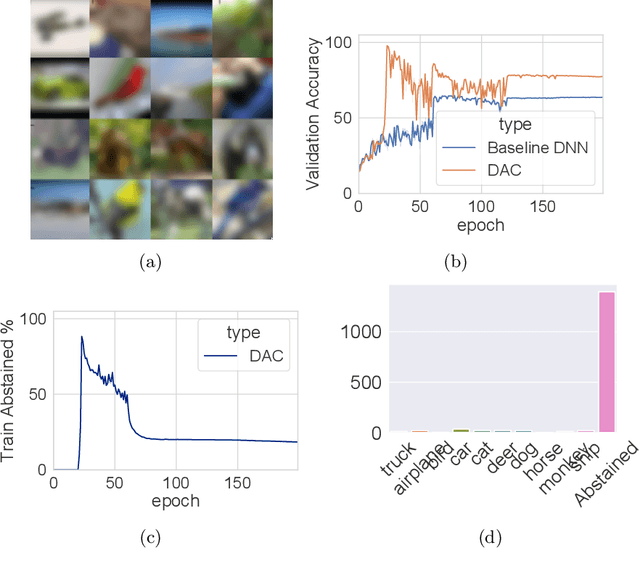
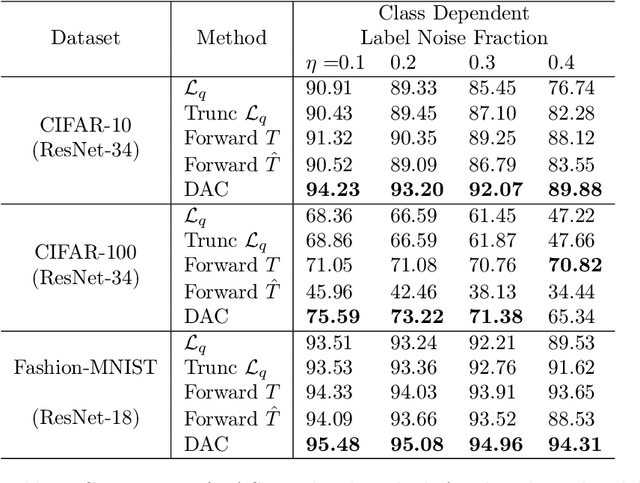
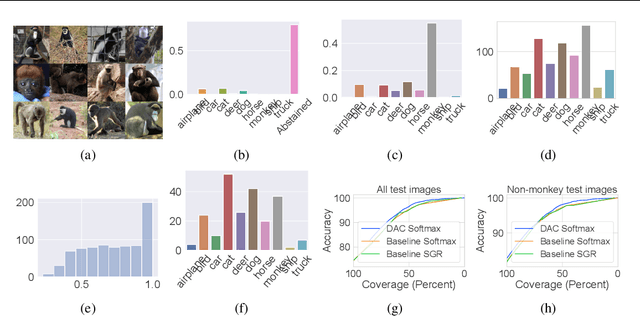
Abstract:We introduce a novel method to combat label noise when training deep neural networks for classification. We propose a loss function that permits abstention during training thereby allowing the DNN to abstain on confusing samples while continuing to learn and improve classification performance on the non-abstained samples. We show how such a deep abstaining classifier (DAC) can be used for robust learning in the presence of different types of label noise. In the case of structured or systematic label noise -- where noisy training labels or confusing examples are correlated with underlying features of the data-- training with abstention enables representation learning for features that are associated with unreliable labels. In the case of unstructured (arbitrary) label noise, abstention during training enables the DAC to be used as an effective data cleaner by identifying samples that are likely to have label noise. We provide analytical results on the loss function behavior that enable dynamic adaption of abstention rates based on learning progress during training. We demonstrate the utility of the deep abstaining classifier for various image classification tasks under different types of label noise; in the case of arbitrary label noise, we show significant improvements over previously published results on multiple image benchmarks.
eAnt-Miner : An Ensemble Ant-Miner to Improve the ACO Classification
Sep 09, 2014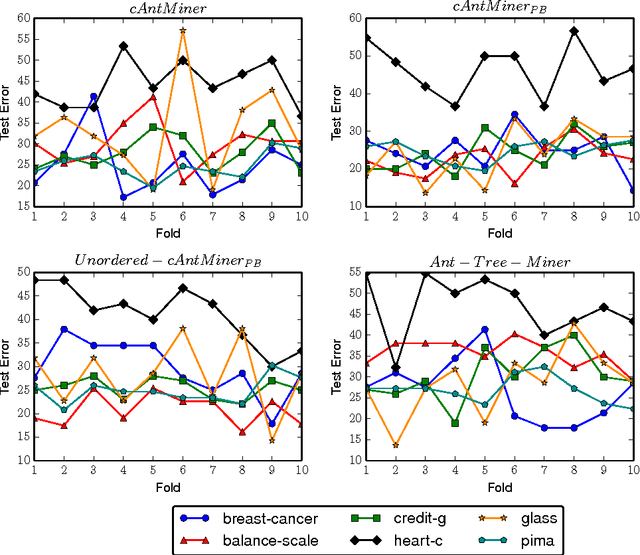
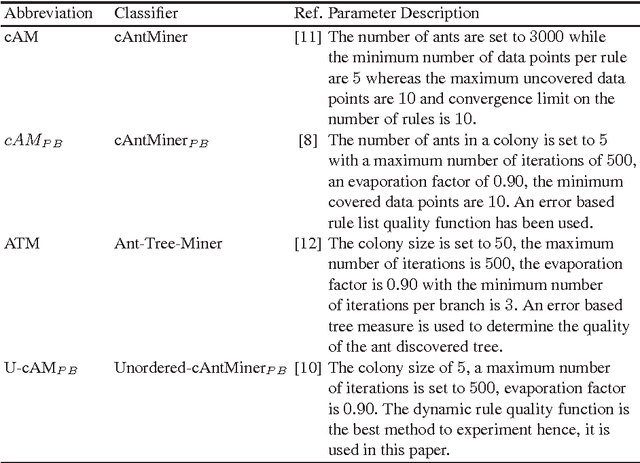
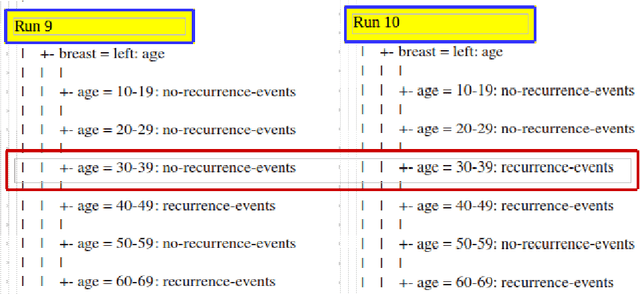
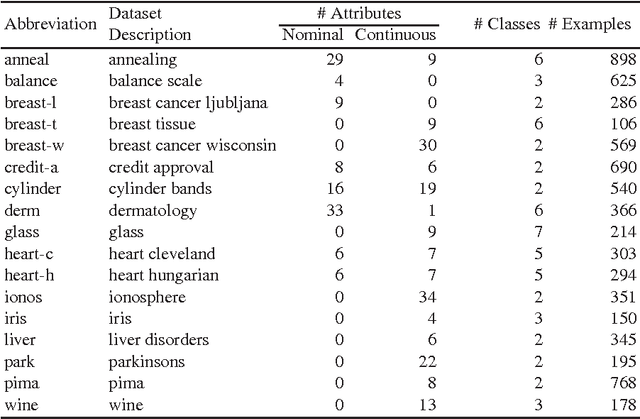
Abstract:Ant Colony Optimization (ACO) has been applied in supervised learning in order to induce classification rules as well as decision trees, named Ant-Miners. Although these are competitive classifiers, the stability of these classifiers is an important concern that owes to their stochastic nature. In this paper, to address this issue, an acclaimed machine learning technique named, ensemble of classifiers is applied, where an ACO classifier is used as a base classifier to prepare the ensemble. The main trade-off is, the predictions in the new approach are determined by discovering a group of models as opposed to the single model classification. In essence, we prepare multiple models from the randomly replaced samples of training data from which, a unique model is prepared by aggregating the models to test the unseen data points. The main objective of this new approach is to increase the stability of the Ant-Miner results there by improving the performance of ACO classification. We found that the ensemble Ant-Miners significantly improved the stability by reducing the classification error on unseen data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge