Farzad Khalvati
Tumor-location-guided CNNs for Pediatric Low-grade Glioma Molecular Biomarker Classification Using MRI
Oct 13, 2022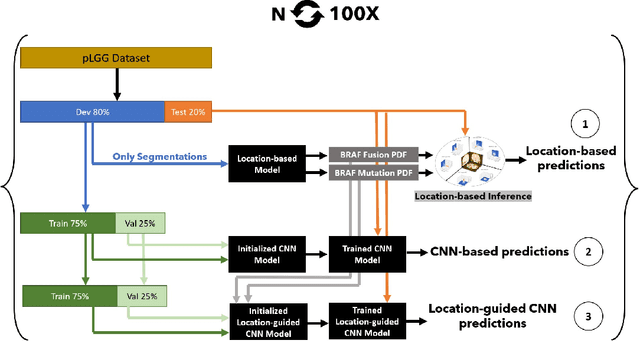
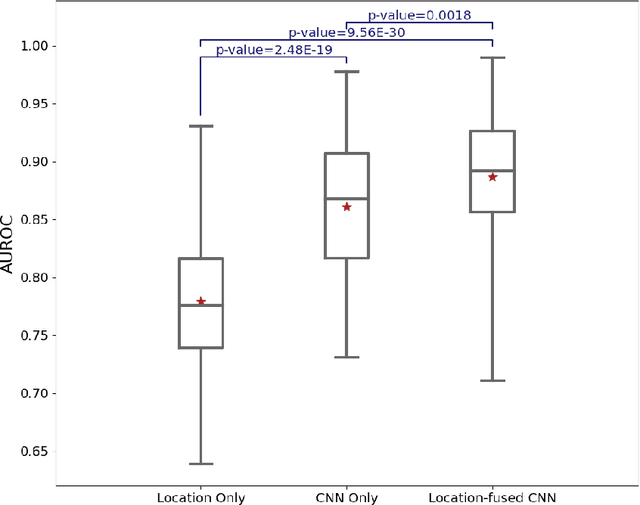
Abstract:Pediatric low-grade glioma (pLGG) is the most common type of brain cancer among children, and the identification of molecular markers for pLGG is crucial for successful treatment planning. Current standard care is biopsy, which is invasive. Thus, the non-invasive imaging-based approaches, where Machine Learning (ML) has a high potential, are impactful. Recently, we developed a tumor-location-based algorithm and demonstrated its potential to differentiate pLGG molecular subtypes. In this work, we first reevaluated the performance of the location-based algorithm on a larger pLGG dataset, which includes 214 patients and achieved an area under the receiver operating characteristic curve (AUROC) of 77.90. A Convolutional Neural Network (CNN) based algorithm increased the average AUROC to 86.11. Ultimately, we designed and implemented a tumor-location-guided CNN algorithm and achieved average AUROC of 88.64. Using a repeated experiment approach with 100 runs, we ensured the results were reproducible and the improvement was statistically significant.
Introducing Vision Transformer for Alzheimer's Disease classification task with 3D input
Oct 03, 2022

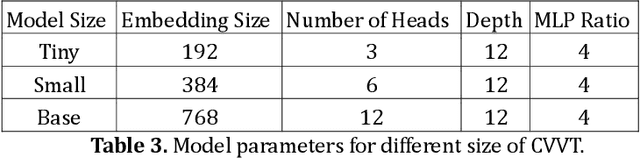
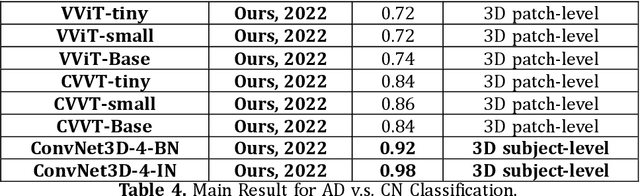
Abstract:Many high-performance classification models utilize complex CNN-based architectures for Alzheimer's Disease classification. We aim to investigate two relevant questions regarding classification of Alzheimer's Disease using MRI: "Do Vision Transformer-based models perform better than CNN-based models?" and "Is it possible to use a shallow 3D CNN-based model to obtain satisfying results?" To achieve these goals, we propose two models that can take in and process 3D MRI scans: Convolutional Voxel Vision Transformer (CVVT) architecture, and ConvNet3D-4, a shallow 4-block 3D CNN-based model. Our results indicate that the shallow 3D CNN-based models are sufficient to achieve good classification results for Alzheimer's Disease using MRI scans.
Superpixel Generation and Clustering for Weakly Supervised Brain Tumor Segmentation in MR Images
Sep 20, 2022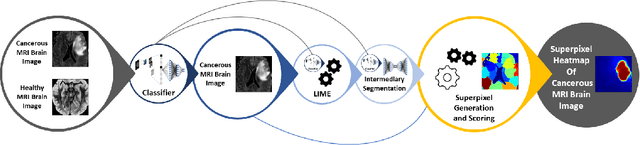
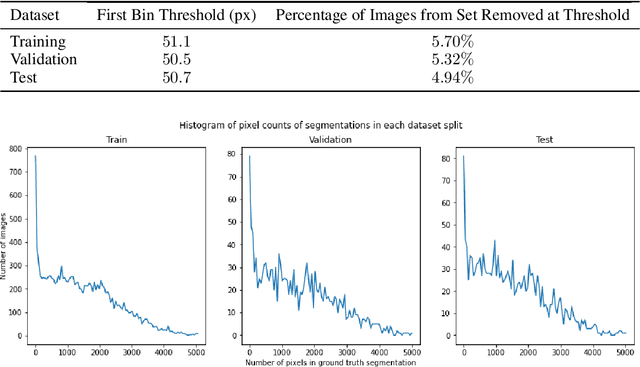
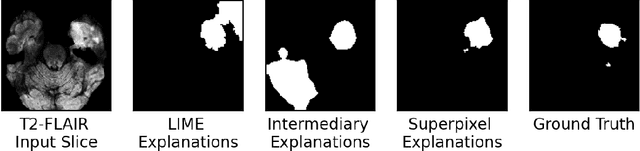
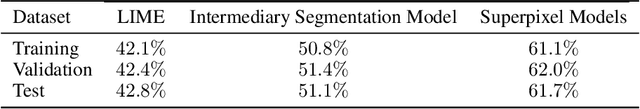
Abstract:Training Machine Learning (ML) models to segment tumors and other anomalies in medical images is an increasingly popular area of research but generally requires manually annotated ground truth segmentations which necessitates significant time and resources to create. This work proposes a pipeline of ML models that utilize binary classification labels, which can be easily acquired, to segment ROIs without requiring ground truth annotations. We used 2D slices of Magnetic Resonance Imaging (MRI) brain scans from the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 dataset and labels indicating the presence of high-grade glioma (HGG) tumors to train the pipeline. Our pipeline also introduces a novel variation of deep learning-based superpixel generation, which enables training guided by clustered superpixels and simultaneously trains a superpixel clustering model. On our test set, our pipeline's segmentations achieved a Dice coefficient of 61.7%, which is a substantial improvement over the 42.8% Dice coefficient acquired when the popular Local Interpretable Model-Agnostic Explanations (LIME) method was used.
Minimizing the Effect of Noise and Limited Dataset Size in Image Classification Using Depth Estimation as an Auxiliary Task with Deep Multitask Learning
Aug 22, 2022
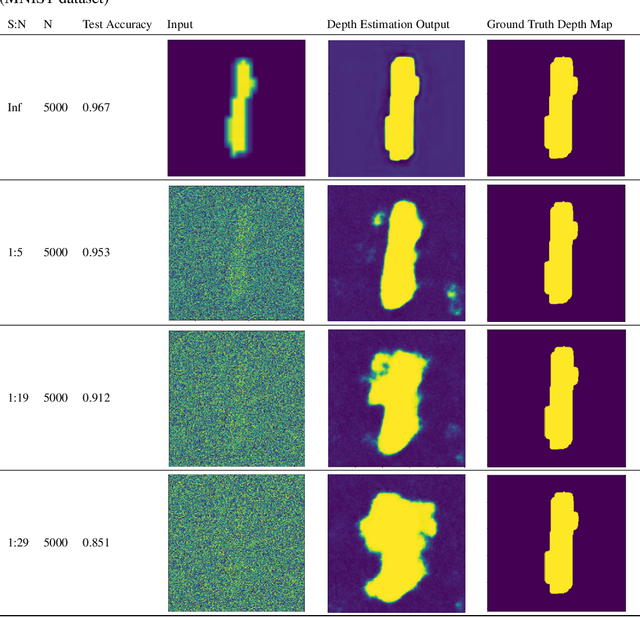

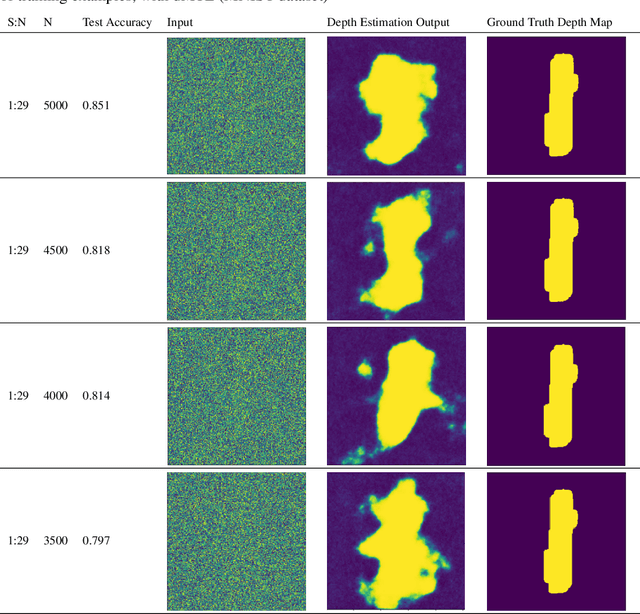
Abstract:Generalizability is the ultimate goal of Machine Learning (ML) image classifiers, for which noise and limited dataset size are among the major concerns. We tackle these challenges through utilizing the framework of deep Multitask Learning (dMTL) and incorporating image depth estimation as an auxiliary task. On a customized and depth-augmented derivation of the MNIST dataset, we show a) multitask loss functions are the most effective approach of implementing dMTL, b) limited dataset size primarily contributes to classification inaccuracy, and c) depth estimation is mostly impacted by noise. In order to further validate the results, we manually labeled the NYU Depth V2 dataset for scene classification tasks. As a contribution to the field, we have made the data in python native format publicly available as an open-source dataset and provided the scene labels. Our experiments on MNIST and NYU-Depth-V2 show dMTL improves generalizability of the classifiers when the dataset is noisy and the number of examples is limited.
Using Multi-modal Data for Improving Generalizability and Explainability of Disease Classification in Radiology
Jul 29, 2022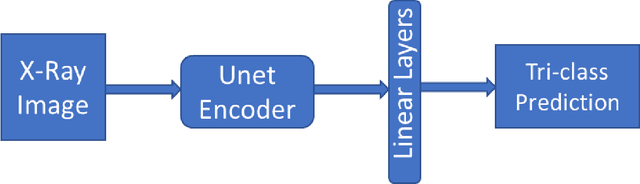

Abstract:Traditional datasets for the radiological diagnosis tend to only provide the radiology image alongside the radiology report. However, radiology reading as performed by radiologists is a complex process, and information such as the radiologist's eye-fixations over the course of the reading has the potential to be an invaluable data source to learn from. Nonetheless, the collection of such data is expensive and time-consuming. This leads to the question of whether such data is worth the investment to collect. This paper utilizes the recently published Eye-Gaze dataset to perform an exhaustive study on the impact on performance and explainability of deep learning (DL) classification in the face of varying levels of input features, namely: radiology images, radiology report text, and radiologist eye-gaze data. We find that the best classification performance of X-ray images is achieved with a combination of radiology report free-text and radiology image, with the eye-gaze data providing no performance boost. Nonetheless, eye-gaze data serving as secondary ground truth alongside the class label results in highly explainable models that generate better attention maps compared to models trained to do classification and attention map generation without eye-gaze data.
Open-radiomics: A Research Protocol to Make Radiomics-based Machine Learning Pipelines Reproducible
Jul 29, 2022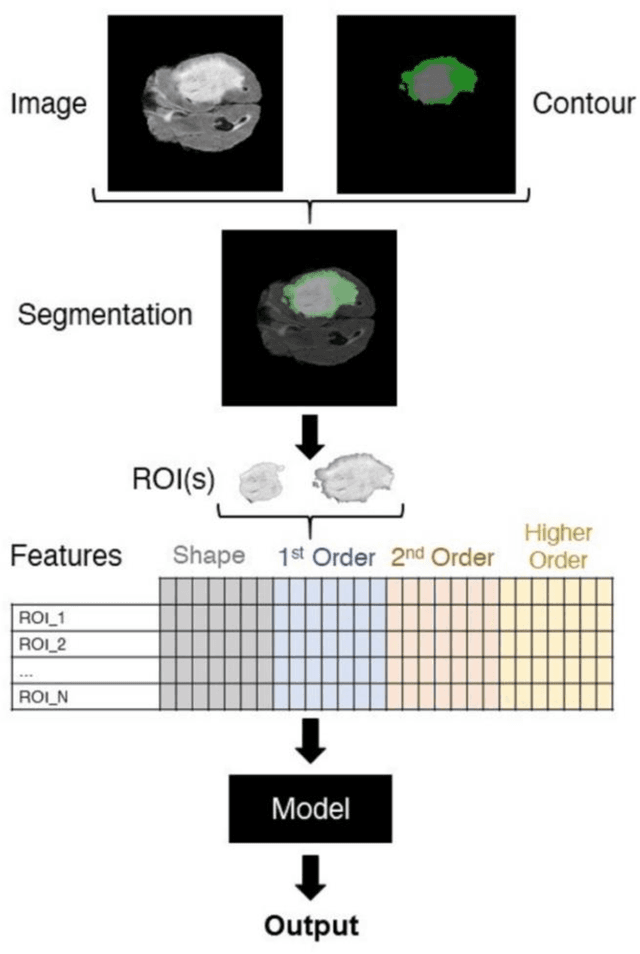
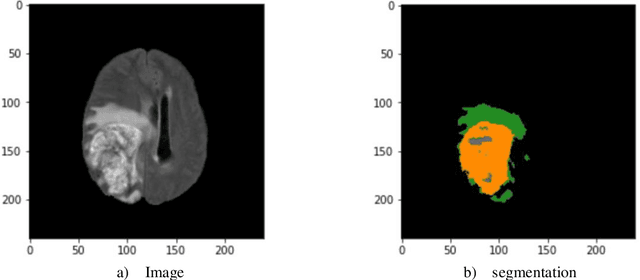
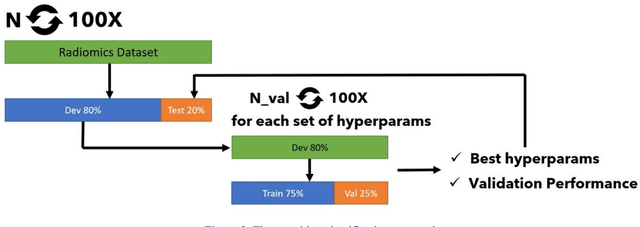
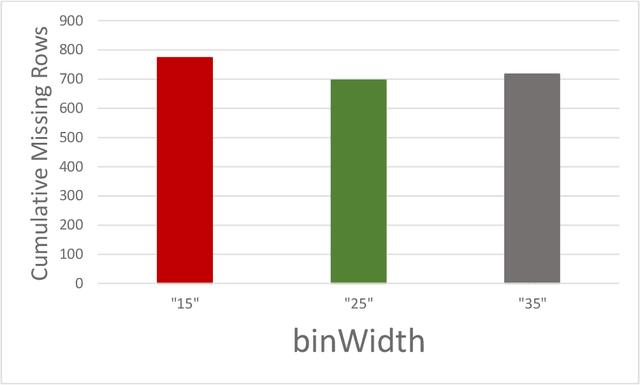
Abstract:The application of artificial intelligence (AI) techniques to medical imaging data has yielded promising results. As an important branch of AI pipelines in medical imaging, radiomics faces two major challenges namely reproducibility and accessibility. In this work, we introduce open-radiomics, a set of radiomics datasets, and a comprehensive radiomics pipeline that investigates the effects of radiomics feature extraction settings such as binWidth and image normalization on the reproducibility of the radiomics results performance. To make radiomics research more accessible and reproducible, we provide guidelines for building machine learning (ML) models on radiomics data, introduce Open-radiomics, an evolving collection of open-source radiomics datasets, and publish baseline models for the datasets.
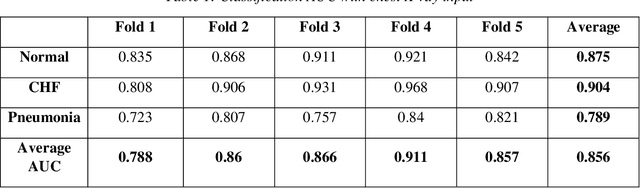
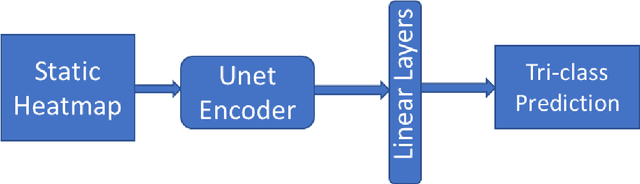
Improving Disease Classification Performance and Explainability of Deep Learning Models in Radiology with Heatmap Generators
Jun 28, 2022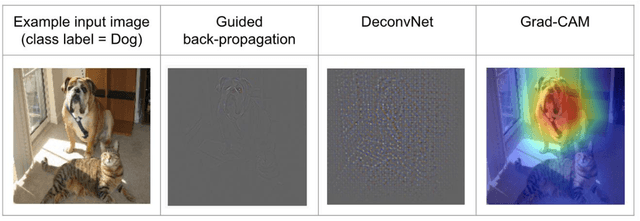

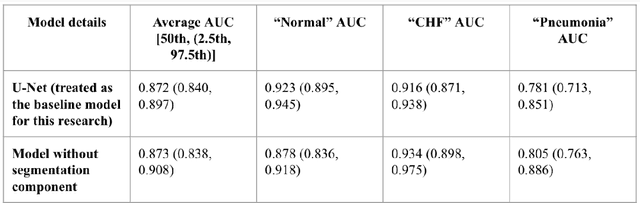
Abstract:As deep learning is widely used in the radiology field, the explainability of such models is increasingly becoming essential to gain clinicians' trust when using the models for diagnosis. In this research, three experiment sets were conducted with a U-Net architecture to improve the classification performance while enhancing the heatmaps corresponding to the model's focus through incorporating heatmap generators during training. All of the experiments used the dataset that contained chest radiographs, associated labels from one of the three conditions ("normal", "congestive heart failure (CHF)", and "pneumonia"), and numerical information regarding a radiologist's eye-gaze coordinates on the images. The paper (A. Karargyris and Moradi, 2021) that introduced this dataset developed a U-Net model, which was treated as the baseline model for this research, to show how the eye-gaze data can be used in multi-modal training for explainability improvement. To compare the classification performances, the 95% confidence intervals (CI) of the area under the receiver operating characteristic curve (AUC) were measured. The best method achieved an AUC of 0.913 (CI: 0.860-0.966). The greatest improvements were for the "pneumonia" and "CHF" classes, which the baseline model struggled most to classify, resulting in AUCs of 0.859 (CI: 0.732-0.957) and 0.962 (CI: 0.933-0.989), respectively. The proposed method's decoder was also able to produce probability masks that highlight the determining image parts in model classifications, similarly as the radiologist's eye-gaze data. Hence, this work showed that incorporating heatmap generators and eye-gaze information into training can simultaneously improve disease classification and provide explainable visuals that align well with how the radiologist viewed the chest radiographs when making diagnosis.
Exploring COVID-19 Related Stressors Using Topic Modeling
Jan 12, 2022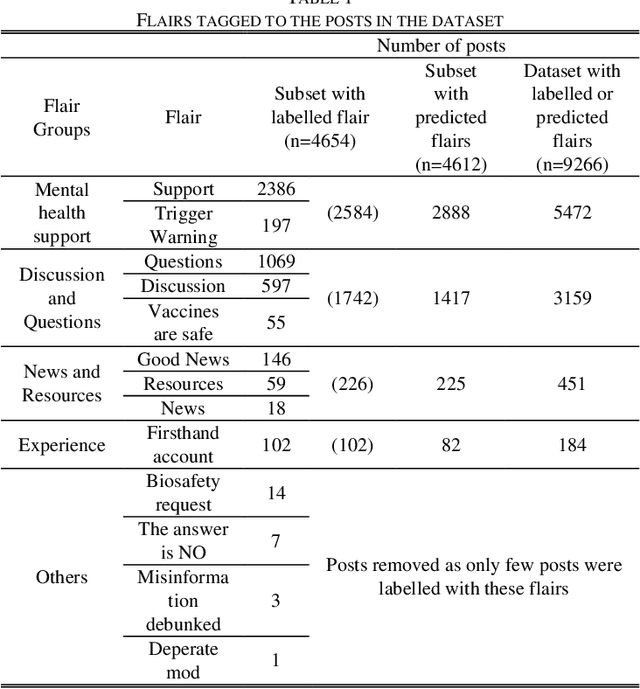
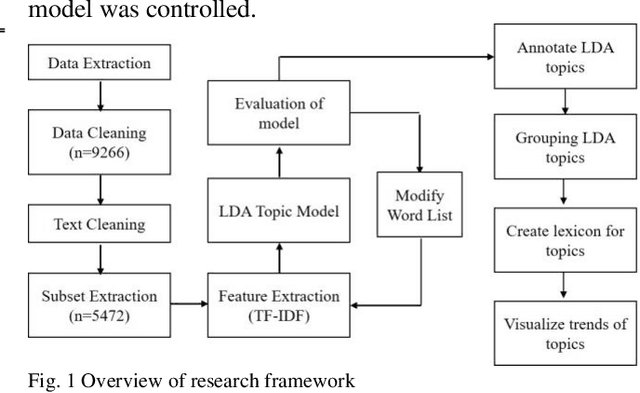

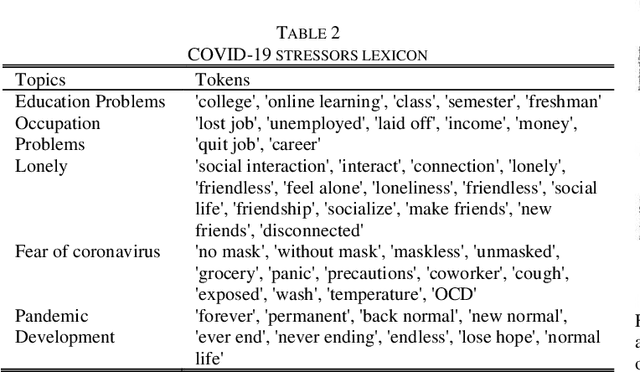
Abstract:The COVID-19 pandemic has affected lives of people from different countries for almost two years. The changes on lifestyles due to the pandemic may cause psychosocial stressors for individuals, and have a potential to lead to mental health problems. To provide high quality mental health supports, healthcare organization need to identify the COVID-19 specific stressors, and notice the trends of prevalence of those stressors. This study aims to apply natural language processing (NLP) on social media data to identify the psychosocial stressors during COVID-19 pandemic, and to analyze the trend on prevalence of stressors at different stages of the pandemic. We obtained dataset of 9266 Reddit posts from subreddit \rCOVID19_support, from 14th Feb ,2020 to 19th July 2021. We used Latent Dirichlet Allocation (LDA) topic model and lexicon methods to identify the topics that were mentioned on the subreddit. Our result presented a dashboard to visualize the trend of prevalence of topics about covid-19 related stressors being discussed on social media platform. The result could provide insights about the prevalence of pandemic related stressors during different stages of COVID-19. The NLP techniques leveraged in this study could also be applied to analyze event specific stressors in the future.
Improving the Segmentation of Pediatric Low-Grade Gliomas through Multitask Learning
Nov 29, 2021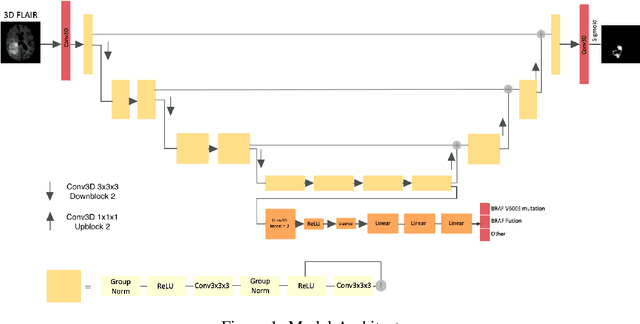
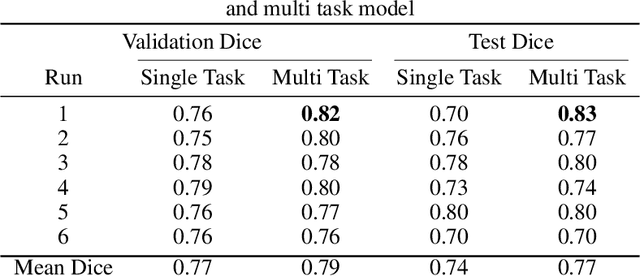
Abstract:Brain tumor segmentation is a critical task for tumor volumetric analyses and AI algorithms. However, it is a time-consuming process and requires neuroradiology expertise. While there has been extensive research focused on optimizing brain tumor segmentation in the adult population, studies on AI guided pediatric tumor segmentation are scarce. Furthermore, MRI signal characteristics of pediatric and adult brain tumors differ, necessitating the development of segmentation algorithms specifically designed for pediatric brain tumors. We developed a segmentation model trained on magnetic resonance imaging (MRI) of pediatric patients with low-grade gliomas (pLGGs) from The Hospital for Sick Children (Toronto, Ontario, Canada). The proposed model utilizes deep Multitask Learning (dMTL) by adding tumor's genetic alteration classifier as an auxiliary task to the main network, ultimately improving the accuracy of the segmentation results.
Localized Perturbations For Weakly-Supervised Segmentation of Glioma Brain Tumours
Nov 29, 2021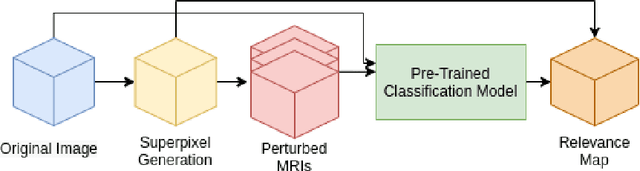
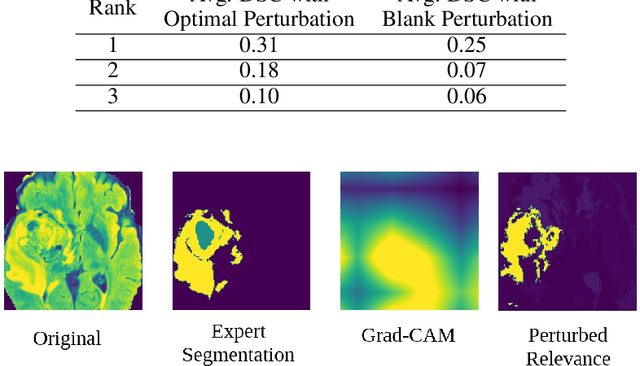
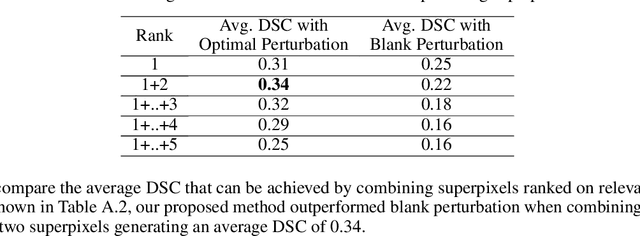

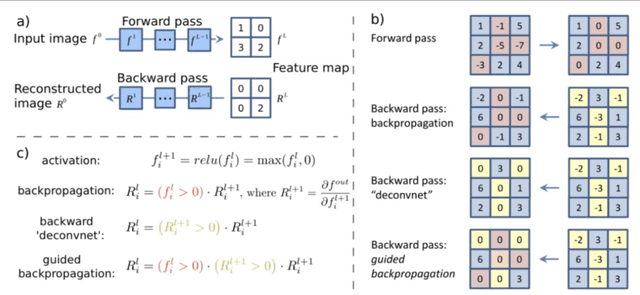
Abstract:Deep convolutional neural networks (CNNs) have become an essential tool in the medical imaging-based computer-aided diagnostic pipeline. However, training accurate and reliable CNNs requires large fine-grain annotated datasets. To alleviate this, weakly-supervised methods can be used to obtain local information from global labels. This work proposes the use of localized perturbations as a weakly-supervised solution to extract segmentation masks of brain tumours from a pretrained 3D classification model. Furthermore, we propose a novel optimal perturbation method that exploits 3D superpixels to find the most relevant area for a given classification using a U-net architecture. Our method achieved a Dice similarity coefficient (DSC) of 0.44 when compared with expert annotations. When compared against Grad-CAM, our method outperformed both in visualization and localization ability of the tumour region, with Grad-CAM only achieving 0.11 average DSC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge